Role of secretory IgA in the mucosal sensing of commensal bacteria
- PMID: 25536286
- PMCID: PMC4615909
- DOI: 10.4161/19490976.2014.983763
Role of secretory IgA in the mucosal sensing of commensal bacteria
Abstract
While the gut epithelium represents the largest mucosal tissue, the mechanisms underlying the interaction between intestinal bacteria and the host epithelium lead to multiple outcomes that remain poorly understood at the molecular level. Deciphering such events may provide valuable information as to the mode of action of commensal and probiotic microorganisms in the gastrointestinal environment. Potential roles of such microorganisms along the privileged target represented by the intestinal immune system include maturation processes prior, during and after weaning, and the reduction of inflammatory reactions in pathogenic conditions. As commensal bacteria are naturally coated by natural and antigen-specific SIgA in the gut lumen, understanding the consequences of such an interaction may provide new clues on how the antibody contributes to homeostasis at mucosal surfaces. This review discusses several aspects of the role of SIgA in the essential communication existing between the host epithelium and members of its microbiota.
Keywords: DC, dendritic cell; IEC, intestinal epithelial cell; SC, secretory component; SED, subepithelial dome; SIgA, secretory IgA; dendritic cells; gut microbiota; intestinal epithelial cells; mucosal homeostasis; pIgR, polymeric Ig receptor; secretory IgA.
Figures

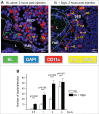
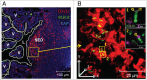
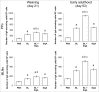
References
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 1977; 31:107-33; PMID:334036; http://dx.doi.org/ 10.1146/annurev.mi.31.100177.000543 - DOI - PubMed
-
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006; 7:688-93; PMID:16819463; http://dx.doi.org/ 10.1038/sj.embor.7400731 - DOI - PMC - PubMed
-
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009; 136:65-80; PMID:19026645; http://dx.doi.org/ 10.1053/j.gastro.2008.10.080 - DOI - PMC - PubMed
-
- Macpherson AJ, Slack E, Geuking MB, McCoy KD. The mucosal firewalls against commensal intestinal microbes. Semin Immunopathol 2009; 31:145-49; PMID:19707762; http://dx.doi.org/ 10.1007/s00281-009-0174-3 - DOI - PubMed
-
- Gutzeit C, Magri G, Cerutti A. Intestinal IgA production and its role in host-microbe interaction. Immunol Rev 2014; 260:76-85; PMID:24942683; http://dx.doi.org/ 10.1111/imr.12189 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
