Sequencing the functional antibody repertoire--diagnostic and therapeutic discovery
- PMID: 25536486
- PMCID: PMC4382308
- DOI: 10.1038/nrrheum.2014.220
Sequencing the functional antibody repertoire--diagnostic and therapeutic discovery
Abstract
The development of high-throughput DNA sequencing technologies has enabled large-scale characterization of functional antibody repertoires, a new method of understanding protective and pathogenic immune responses. Important parameters to consider when sequencing antibody repertoires include the methodology, the B-cell population and clinical characteristics of the individuals analysed, and the bioinformatic analysis. Although focused sequencing of immunoglobulin heavy chains or complement determining regions can be utilized to monitor particular immune responses and B-cell malignancies, high-fidelity analysis of the full-length paired heavy and light chains expressed by individual B cells is critical for characterizing functional antibody repertoires. Bioinformatic identification of clonal antibody families and recombinant expression of representative members produces recombinant antibodies that can be used to identify the antigen targets of functional immune responses and to investigate the mechanisms of their protective or pathogenic functions. Integrated analysis of coexpressed functional genes provides the potential to further pinpoint the most important antibodies and clonal families generated during an immune response. Sequencing antibody repertoires is transforming our understanding of immune responses to autoimmunity, vaccination, infection and cancer. We anticipate that antibody repertoire sequencing will provide next-generation biomarkers, diagnostic tools and therapeutic antibodies for a spectrum of diseases, including rheumatic diseases.
Figures
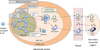


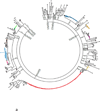
References
-
- Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. - PubMed
-
- Rosen A, Casciola-Rosen L. Autoantigens as substrates for apoptotic proteases: implications for the pathogenesis of systemic autoimmune disease. Cell Death Differ. 1999;6:6–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

