Impact of centralization on aCGH-based genomic profiles for precision medicine in oncology
- PMID: 25538175
- PMCID: PMC4343013
- DOI: 10.1093/annonc/mdu582
Impact of centralization on aCGH-based genomic profiles for precision medicine in oncology
Abstract
Background: Comparative genomic hybridization (CGH) arrays are increasingly used in personalized medicine programs to identify gene copy number aberrations (CNAs) that may be used to guide clinical decisions made during molecular tumor boards. However, analytical processes such as the centralization step may profoundly affect CGH array results and therefore may adversely affect outcomes in the precision medicine context.
Patients and methods: The effect of three different centralization methods: median, maximum peak, alternative peak, were evaluated on three datasets: (i) the NCI60 cell lines panel, (ii) the Cancer Cell Line Encyclopedia (CCLE) panel, and (iii) the patients enrolled in prospective molecular screening trials (SAFIR-01 n = 283, MOSCATO-01 n = 309), and compared with karyotyping, drug sensitivity, and patient-drug matching, respectively.
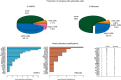
Results: Using the NCI60 cell lines panel, the profiles generated by the alternative peak method were significantly closer to the cell karyotypes than those generated by the other centralization strategies (P < 0.05). Using the CCLE dataset, selected genes (ERBB2, EGFR) were better or equally correlated to the IC50 of their companion drug (lapatinib, erlotinib), when applying the alternative centralization. Finally, focusing on 24 actionable genes, we observed as many as 7.1% (SAFIR-01) and 6.8% (MOSCATO-01) of patients originally not oriented to a specific treatment, but who could have been proposed a treatment based on the alternative peak centralization method.
Conclusion: The centralization method substantially affects the call detection of CGH profiles and may thus impact precision medicine approaches. Among the three methods described, the alternative peak method addresses limitations associated with existing approaches.
Keywords: aCGH; comparative genomic hybridization; precision medicine; targeted therapy.
© The Author 2014. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


References
-
- André F, Bachelot T, Commo F, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. - PubMed
-
- Hollebecque A, Massard C, De Baere T, et al. Molecular screening for cancer treatment optimization (MOSCATO 01): a prospective molecular triage trial—interim results. ASCO Annual Meeting. 2013 Abstract 2512.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

