Mass spectrometry-based detection and assignment of protein posttranslational modifications
- PMID: 25541750
- PMCID: PMC4301092
- DOI: 10.1021/cb500904b
Mass spectrometry-based detection and assignment of protein posttranslational modifications
Abstract
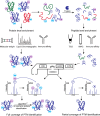
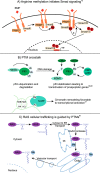
Recent advances in mass spectrometry (MS)-based proteomics allow the identification and quantitation of thousands of posttranslational modification (PTM) sites in a single experiment. This follows from the development of more effective class enrichment strategies, new high performance instrumentation and bioinformatic algorithms with rigorous scoring strategies. More widespread use of these combined capabilities have led to a vast expansion in our knowledge of the complexity of biological processes mediated by PTMs. The classes most actively pursued include phosphorylation, ubiquitination, O-GlcNAcylation, methylation, and acetylation. Very recently succinylation, SUMOylation, and citrullination have emerged. Among the some 260 000 PTM sites that have been identified in the human proteome thus far, only a few have been assigned to key regulatory and/or other biological roles. Here, we provide an update of MS-based PTM analyses, with a focus on current enrichment strategies coupled with revolutionary advances in high performance MS. Furthermore, we discuss examples of the discovery of recently described biological roles of PTMs and address the challenges of defining site-specific functions.
Figures
Similar articles
-
Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics.Adv Exp Med Biol. 2016;919:345-382. doi: 10.1007/978-3-319-41448-5_17. Adv Exp Med Biol. 2016. PMID: 27975226 Review.
-
Advances in purification and separation of posttranslationally modified proteins.J Proteomics. 2013 Oct 30;92:2-27. doi: 10.1016/j.jprot.2013.05.040. Epub 2013 Jun 15. J Proteomics. 2013. PMID: 23777897 Review.
-
Mass Spectrometry Analysis of Lysine Posttranslational Modifications of Tau Protein from Alzheimer's Disease Brain.Methods Mol Biol. 2017;1523:161-177. doi: 10.1007/978-1-4939-6598-4_10. Methods Mol Biol. 2017. PMID: 27975250 Free PMC article.
-
The next level of complexity: crosstalk of posttranslational modifications.Proteomics. 2014 Mar;14(4-5):513-24. doi: 10.1002/pmic.201300344. Epub 2014 Jan 6. Proteomics. 2014. PMID: 24339426 Review.
-
Functional Proteomic Analysis to Characterize Signaling Crosstalk.Methods Mol Biol. 2019;1871:197-224. doi: 10.1007/978-1-4939-8814-3_14. Methods Mol Biol. 2019. PMID: 30276742
Cited by
-
In-depth analysis of the Sirtuin 5-regulated mouse brain malonylome and succinylome using library-free data-independent acquisitions.Proteomics. 2023 Feb;23(3-4):e2100371. doi: 10.1002/pmic.202100371. Epub 2022 Dec 18. Proteomics. 2023. PMID: 36479818 Free PMC article.
-
Control of protein stability by post-translational modifications.Nat Commun. 2023 Jan 13;14(1):201. doi: 10.1038/s41467-023-35795-8. Nat Commun. 2023. PMID: 36639369 Free PMC article. Review.
-
Rapid and In-Depth Coverage of the (Phospho-)Proteome With Deep Libraries and Optimal Window Design for dia-PASEF.Mol Cell Proteomics. 2022 Sep;21(9):100279. doi: 10.1016/j.mcpro.2022.100279. Epub 2022 Aug 6. Mol Cell Proteomics. 2022. PMID: 35944843 Free PMC article.
-
Robust unsupervised deconvolution of linear motifs characterizes 68 protein modifications at proteome scale.Sci Rep. 2021 Nov 18;11(1):22490. doi: 10.1038/s41598-021-01971-3. Sci Rep. 2021. PMID: 34795380 Free PMC article.
-
Characterization of Prenylated C-terminal Peptides Using a Thiopropyl-based Capture Technique and LC-MS/MS.Mol Cell Proteomics. 2020 Jun;19(6):1005-1016. doi: 10.1074/mcp.RA120.001944. Epub 2020 Apr 13. Mol Cell Proteomics. 2020. PMID: 32284353 Free PMC article.
References
-
- Consortium I. H. G. S. (2004) Finishing the euchromatic sequence of the human genome. Nature 431, 931–945. - PubMed
-
- Jensen O. N. (2004) Modification-specific proteomics: Characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8, 33–41. - PubMed
-
- Walsh C. T. (2006) Posttranslational Modification of Proteins: Expanding Nature’s Inventory, 1st ed., pp 1–490, Roberts and Company, Publishers, Greenwood Village, CO.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

