Adaptive combination of P-values for family-based association testing with sequence data
- PMID: 25541952
- PMCID: PMC4277421
- DOI: 10.1371/journal.pone.0115971
Adaptive combination of P-values for family-based association testing with sequence data
Abstract
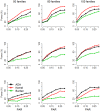
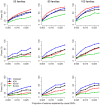
Family-based study design will play a key role in identifying rare causal variants, because rare causal variants can be enriched in families with multiple affected subjects. Furthermore, different from population-based studies, family studies are robust to bias induced by population substructure. It is well known that rare causal variants are difficult to detect from single-locus tests. Therefore, burden tests and non-burden tests have been developed, by combining signals of multiple variants in a chromosomal region or a functional unit. This inevitably incorporates some neutral variants into the test statistics, which can dilute the power of statistical methods. To guard against the noise caused by neutral variants, we here propose an 'adaptive combination of P-values method' (abbreviated as 'ADA'). This method combines per-site P-values of variants that are more likely to be causal. Variants with large P-values (which are more likely to be neutral variants) are discarded from the combined statistic. In addition to performing extensive simulation studies, we applied these tests to the Genetic Analysis Workshop 17 data sets, where real sequence data were generated according to the 1000 Genomes Project. Compared with some existing methods, ADA is more robust to the inclusion of neutral variants. This is a merit especially when dichotomous traits are analyzed. However, there are some limitations for ADA. First, it is more computationally intensive. Second, pedigree structures and founders' sequence data are required for the permutation procedure. Third, unrelated controls cannot be included. We here show that, for family-based studies, the application of ADA is limited to dichotomous trait analyses with full pedigree information.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

