SIRT6 minor allele genotype is associated with >5-year decrease in lifespan in an aged cohort
- PMID: 25541994
- PMCID: PMC4277407
- DOI: 10.1371/journal.pone.0115616
SIRT6 minor allele genotype is associated with >5-year decrease in lifespan in an aged cohort
Abstract
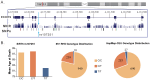
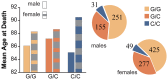
Aging is a natural process involving complex interplay between environment, metabolism, and genes. Sirtuin genes and their downstream targets have been associated with lifespan in numerous organisms from nematodes to humans. Several target proteins of the sirtuin genes are key sensors and/or effectors of oxidative stress pathways including FOXO3, SOD3, and AKT1. To examine the relationship between single nucleotide polymorphisms (SNP) at candidate genes in these pathways and human lifespan, we performed a molecular epidemiologic study of an elderly cohort (≥65 years old.). Using age at death as a continuous outcome variable and assuming a co-dominant genetic model within the framework of multi-variable linear regression analysis, the genotype-specific adjusted mean age at death was estimated for individual SNP genotypes while controlling for age-related risk factors including smoking, body mass index, alcohol consumption and co-morbidity. Significant associations were detected between human lifespan and SNPs in genes SIRT3, SIRT5, SIRT6, FOXO3 and SOD3. Individuals with either the CC or CT genotype at rs107251 within SIRT6 displayed >5-year mean survival advantages compared to the TT genotype (5.5 and 5.9 years, respectively; q-value = 0.012). Other SNPs revealed genotype-specific mean survival advantages ranging from 0.5 to 1.6 years. Gender also modified the effect of SNPs in SIRT3, SIRT5 and AKT1 on lifespan. Our novel findings highlight the impact of sirtuins and sirtuin-related genotypes on lifespan, the importance of evaluating gender and the advantage of using age as a continuous variable in analyses to report mean age at death.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

