Analysis of potato virus Y coat protein epitopes recognized by three commercial monoclonal antibodies
- PMID: 25542005
- PMCID: PMC4277358
- DOI: 10.1371/journal.pone.0115766
Analysis of potato virus Y coat protein epitopes recognized by three commercial monoclonal antibodies
Abstract
Background: Potato virus Y (PVY, genus Potyvirus) causes substantial economic losses in solanaceous plants. Routine screening for PVY is an essential part of seed potato certification, and serological assays are often used. The commercial, commonly used monoclonal antibodies, MAb1128, MAb1129, and MAb1130, recognize the viral coat protein (CP) of PVY and distinguish PVYN strains from PVYO and PVYC strains, or detect all PVY strains, respectively. However, the minimal epitopes recognized by these antibodies have not been identified.
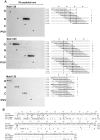
Methodology/principal findings: SPOT peptide array was used to map the epitopes in CP recognized by MAb1128, MAb1129, and MAb1130. Then alanine replacement as well as N- and C-terminal deletion analysis of the identified peptide epitopes was done to determine critical amino acids for antibody recognition and the respective minimal epitopes. The epitopes of all antibodies were located within the 30 N-terminal-most residues. The minimal epitope of MAb1128 was 25NLNKEK30. Replacement of 25N or 27N with alanine weakened the recognition by MAb1128, and replacement of 26L, 29E, or 30K nearly precluded recognition. The minimal epitope for MAb1129 was 16RPEQGSIQSNP26 and the most critical residues for recognition were 22I and 23Q. The epitope of MAb1130 was defined by residues 5IDAGGS10. Mutation of residue 6D abrogated and mutation of 9G strongly reduced recognition of the peptide by MAb1130. Amino acid sequence alignment demonstrated that these epitopes are relatively conserved among PVY strains. Finally, recombinant CPs were produced to demonstrate that mutations in the variable positions of the epitope regions can affect detection with the MAbs.
Conclusions/significance: The epitope data acquired can be compared with data on PVY CP-encoding sequences produced by laboratories worldwide and utilized to monitor how widely the new variants of PVY can be detected with current seed potato certification schemes or during the inspection of imported seed potatoes as conducted with these MAbs.
Conflict of interest statement
Figures



References
-
- Adams MJ, Zerbini FM, French R, Rabenstein F, Stenger DC, et al. (2012) Family Potyviridae. In: King AM, Adams MJ, Carstens EB and Lefkowich EJeditors. Virus Taxonomy. San Diego: Elsevier, pp. 1069–1089.
-
- Valkonen JPT (2007) Chapter 28 - Viruses: Economical losses and biotechnological potential. In: Vreugdenhil D, Bradshaw J, GebhardtC, Govers F, Mackerron DKL, Taylor M Aet al. editors. Potato Biology and Biotechnology. Amsterdam: Elsevier Science B.V. pp. 619–641.
-
- Tian YP, Liu JL, Zhang CL, Liu YY, Wang B, et al. (2011) Genetic diversity of Potato virus Y infecting tobacco crops in China. Phytopathology 101:377–387. - PubMed
-
- Romero A, Blanco-Urgoiti B, Soto MJ, Fereres A, Ponz F (2001) Characterization of typical pepper-isolates of PVY reveals multiple pathotypes within a single genetic strain. Virus Res 79:71–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

