Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex
- PMID: 25543282
- PMCID: PMC4293355
- DOI: 10.1016/j.devcel.2014.10.027
Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex
Abstract
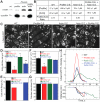
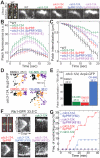
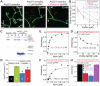
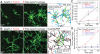
Fission yeast cells use Arp2/3 complex and formin to assemble diverse filamentous actin (F-actin) networks within a common cytoplasm for endocytosis, division, and polarization. Although these homeostatic F-actin networks are usually investigated separately, competition for a limited pool of actin monomers (G-actin) helps to regulate their size and density. However, the mechanism by which G-actin is correctly distributed between rival F-actin networks is not clear. Using a combination of cell biological approaches and in vitro reconstitution of competition between actin assembly factors, we found that the small G-actin binding protein profilin directly inhibits Arp2/3 complex-mediated actin assembly. Profilin is therefore required for formin to compete effectively with excess Arp2/3 complex for limited G-actin and to assemble F-actin for contractile ring formation in dividing cells.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Global resource distribution: allocation of actin building blocks by profilin.Dev Cell. 2015 Jan 12;32(1):5-6. doi: 10.1016/j.devcel.2014.12.022. Dev Cell. 2015. PMID: 25584793
-
Competition and collaboration between different actin assembly pathways allows for homeostatic control of the actin cytoskeleton.Bioarchitecture. 2014;5(1-2):27-34. doi: 10.1080/19490992.2015.1090670. Epub 2015 Oct 2. Bioarchitecture. 2014. PMID: 26430713 Free PMC article.
References
-
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. - PubMed
-
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

