The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer
- PMID: 25544564
- PMCID: PMC4764128
- DOI: 10.1016/j.molcel.2014.11.021
The strength and cooperativity of KIT ectodomain contacts determine normal ligand-dependent stimulation or oncogenic activation in cancer
Abstract
The receptor tyrosine kinase KIT plays an important role in development of germ cells, hematopoietic cells, and interstitial pacemaker cells. Oncogenic KIT mutations play an important "driver" role in gastrointestinal stromal tumors, acute myeloid leukemias, and melanoma, among other cancers. Here we describe the crystal structure of a recurring somatic oncogenic mutation located in the C-terminal Ig-like domain (D5) of the ectodomain, rendering KIT tyrosine kinase activity constitutively activated. The structural analysis, together with biochemical and biophysical experiments and detailed analyses of the activities of a variety of oncogenic KIT mutations, reveals that the strength of homotypic contacts and the cooperativity in the action of D4D5 regions determines whether KIT is normally regulated or constitutively activated in cancers. We propose that cooperative interactions mediated by multiple weak homotypic contacts between receptor molecules are responsible for regulating normal ligand-dependent or oncogenic RTK activation via a "zipper-like" mechanism for receptor activation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
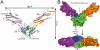





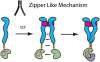
References
-
- Anderson DM, Williams DE, Tushinski R, Gimpel S, Eisenman J, Cannizzaro LA, Aronson M, Croce CM, Huebner K, Cosman D. Alternate splicing of mRNAs encoding human mast cell growth factor and localization of the gene to chromosome 12q22-q24. Cell Growth Differ. 1991;2:373–378. - PubMed
-
- Ashman LK. The biology of stem cell factor and its receptor C-kit. Int. J. Biochem. Cell Biol. 1999;31:1037–1051. - PubMed
-
- Ashman LK, Griffith R. Therapeutic targeting of c-KIT in cancer. Expert Opin. Investig. Drugs. 2012:1–13. - PubMed
-
- Bae JH, Schlessinger J. Asymmetric tyrosine kinase arrangements in activation or autophosphorylation of receptor tyrosine kinases. Mol. Cells. 2010;29:443–448. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

