Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations
- PMID: 25547679
- PMCID: PMC4359967
- DOI: 10.1158/1078-0432.CCR-14-2146
Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations
Abstract
Purpose: Although tyrosine kinase inhibitors (TKI) can be effective therapies for leukemia, they fail to fully eliminate leukemic cells and achieve durable remissions for many patients with advanced BCR-ABL(+) leukemias or acute myelogenous leukemia (AML). Through a large-scale synthetic lethal RNAi screen, we identified pyruvate dehydrogenase, the limiting enzyme for pyruvate entry into the mitochondrial tricarboxylic acid cycle, as critical for the survival of chronic myelogenous leukemia (CML) cells upon BCR-ABL inhibition. Here, we examined the role of mitochondrial metabolism in the survival of Ph(+) leukemia and AML upon TK inhibition.
Experimental design: Ph(+) cancer cell lines, AML cell lines, leukemia xenografts, cord blood, and patient samples were examined.
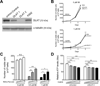
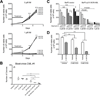
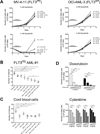
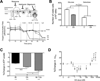
Results: We showed that the mitochondrial ATP-synthase inhibitor oligomycin-A greatly sensitized leukemia cells to TKI in vitro. Surprisingly, oligomycin-A sensitized leukemia cells to BCR-ABL inhibition at concentrations of 100- to 1,000-fold below those required for inhibition of respiration. Oligomycin-A treatment rapidly led to mitochondrial membrane depolarization and reduced ATP levels, and promoted superoxide production and leukemia cell apoptosis when combined with TKI. Importantly, oligomycin-A enhanced elimination of BCR-ABL(+) leukemia cells by TKI in a mouse model and in primary blast crisis CML samples. Moreover, oligomycin-A also greatly potentiated the elimination of FLT3-dependent AML cells when combined with an FLT3 TKI, both in vitro and in vivo.
Conclusions: TKI therapy in leukemia cells creates a novel metabolic state that is highly sensitive to particular mitochondrial perturbations. Targeting mitochondrial metabolism as an adjuvant therapy could therefore improve therapeutic responses to TKI for patients with BCR-ABL(+) and FLT3(ITD) leukemias.
©2014 American Association for Cancer Research.
Conflict of interest statement
Figures

References
-
- Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. - PubMed
-
- Foa R, Vitale A, Vignetti M, Meloni G, Guarini A, De Propris MS, et al. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;118:6521–6528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 RR025780/RR/NCRR NIH HHS/United States
- K01 CA133182/CA/NCI NIH HHS/United States
- F31-CA157166/CA/NCI NIH HHS/United States
- T32 GM008497/GM/NIGMS NIH HHS/United States
- T32 CA082086/CA/NCI NIH HHS/United States
- K22-CA172757/CA/NCI NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- T32 CA174648/CA/NCI NIH HHS/United States
- P30-CA046934/CA/NCI NIH HHS/United States
- K01-CA133182/CA/NCI NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- 5UL1-RR025780/RR/NCRR NIH HHS/United States
- K22 CA172757/CA/NCI NIH HHS/United States
- F31 CA157166/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

