The GRAS gene family in pine: transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots
- PMID: 25547982
- PMCID: PMC4302573
- DOI: 10.1186/s12870-014-0354-8
The GRAS gene family in pine: transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots
Abstract
Background: Adventitious rooting is an organogenic process by which roots are induced from differentiated cells other than those specified to develop roots. In forest tree species, age and maturation are barriers to adventitious root formation by stem cuttings. The mechanisms behind the respecification of fully differentiated progenitor cells, which underlies adventitious root formation, are unknown.
Results: Here, the GRAS gene family in pine is characterized and the expression of a subset of these genes during adventitious rooting is reported. Comparative analyses of protein structures showed that pine GRAS members are conserved compared with their relatives in angiosperms. Relatively high GRAS mRNA levels were measured in non-differentiated proliferating embryogenic cultures and during embryo development. The mRNA levels of putative GRAS family transcription factors, including Pinus radiata's SCARECROW (SCR), PrSCR, and SCARECROW-LIKE (SCL) 6, PrSCL6, were significantly reduced or non-existent in adult tissues that no longer had the capacity to form adventitious roots, but were maintained or induced after the reprogramming of adult cells in rooting-competent tissues. A subset of genes, SHORT-ROOT (PrSHR), PrSCL1, PrSCL2, PrSCL10 and PrSCL12, was also expressed in an auxin-, age- or developmental-dependent manner during adventitious root formation.
Conclusions: The GRAS family of pine has been characterized by analyzing protein structures, phylogenetic relationships, conserved motifs and gene expression patterns. Individual genes within each group have acquired different and specialized functions, some of which could be related to the competence and reprogramming of adult cells to form adventitious roots.
Figures
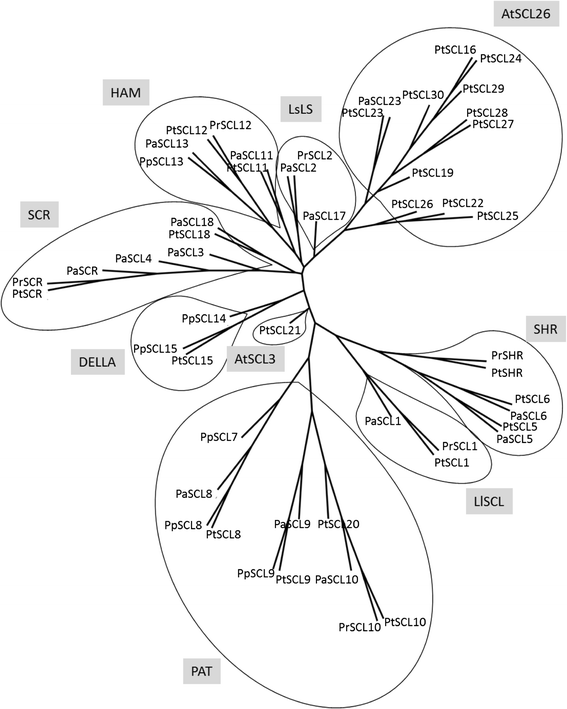
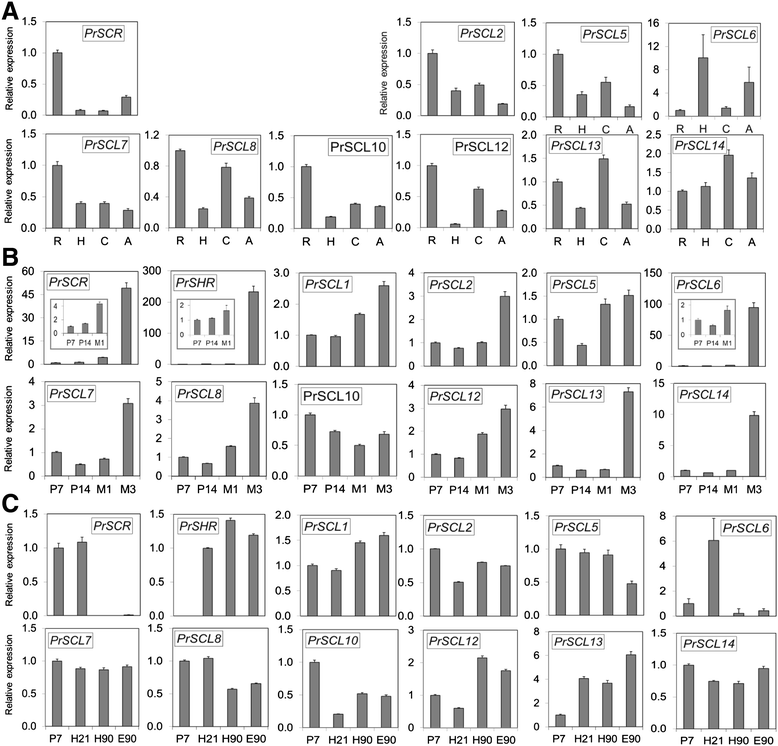
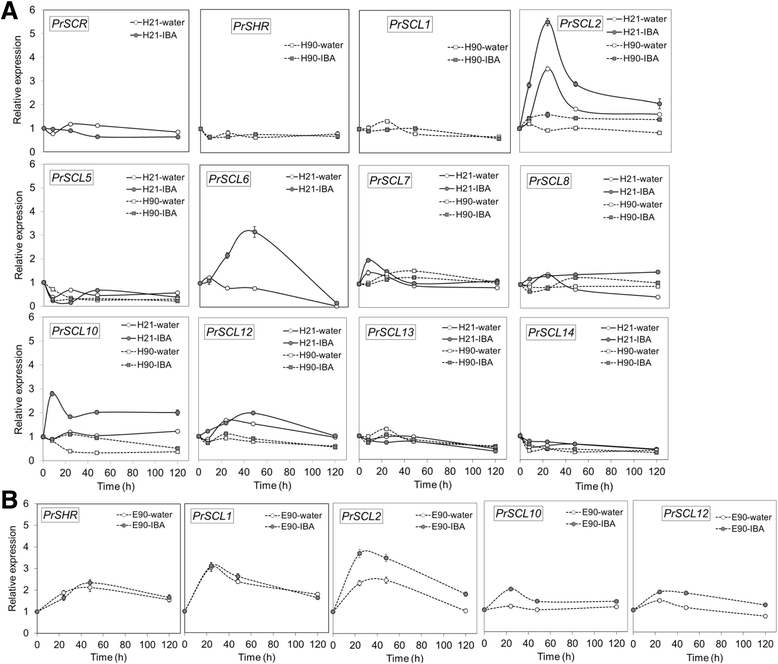
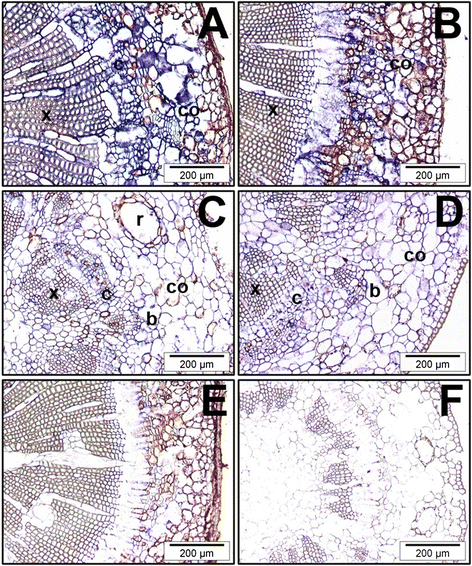
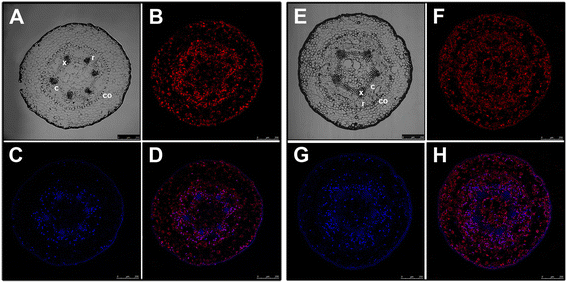
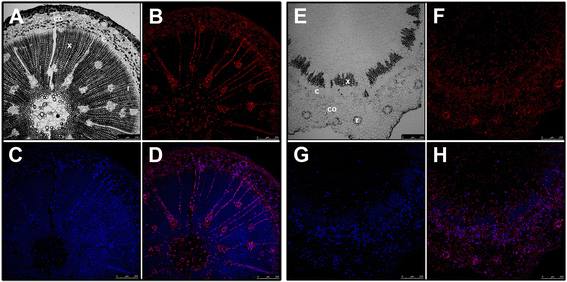
References
-
- Greenwood MS. Rejuvenation of forest trees. Plant Growth Regul. 1987;6:1–12. doi: 10.1007/BF00043947. - DOI
-
- Greenwood MS, Hutchison KW. Maturation as a developmental process. In: Ahuja MR, Libby WJ, editors. Clonal Forestry: Genetics, Biotechnology and Application. New York: Springer Verlag; 1993. pp. 14–33.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

