Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense
- PMID: 25548049
- PMCID: PMC4325140
- DOI: 10.1128/AEM.03795-14
Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense
Abstract
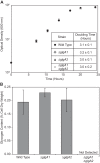
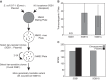
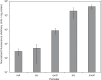
Aerobic methanotrophs oxidize methane at ambient temperatures and pressures and are therefore attractive systems for methane-based bioconversions. In this work, we developed and validated genetic tools for Methylomicrobium buryatense, a haloalkaliphilic gammaproteobacterial (type I) methanotroph. M. buryatense was isolated directly on natural gas and grows robustly in pure culture with a 3-h doubling time, enabling rapid genetic manipulation compared to many other methanotrophic species. As a proof of concept, we used a sucrose counterselection system to eliminate glycogen production in M. buryatense by constructing unmarked deletions in two redundant glycogen synthase genes. We also selected for a more genetically tractable variant strain that can be conjugated with small incompatibility group P (IncP)-based broad-host-range vectors and determined that this capability is due to loss of the native plasmid. These tools make M. buryatense a promising model system for studying aerobic methanotroph physiology and enable metabolic engineering in this bacterium for industrial biocatalysis of methane.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Electroporation-Based Genetic Manipulation in Type I Methanotrophs.Appl Environ Microbiol. 2016 Jan 22;82(7):2062-2069. doi: 10.1128/AEM.03724-15. Appl Environ Microbiol. 2016. PMID: 26801578 Free PMC article.
-
A modular approach for high-flux lactic acid production from methane in an industrial medium using engineered Methylomicrobium buryatense 5GB1.J Ind Microbiol Biotechnol. 2018 Jun;45(6):379-391. doi: 10.1007/s10295-018-2035-3. Epub 2018 Apr 19. J Ind Microbiol Biotechnol. 2018. PMID: 29675615
-
Core Metabolism Shifts during Growth on Methanol versus Methane in the Methanotroph Methylomicrobium buryatense 5GB1.mBio. 2019 Apr 9;10(2):e00406-19. doi: 10.1128/mBio.00406-19. mBio. 2019. PMID: 30967465 Free PMC article.
-
Extrachromosomal, extraordinary and essential--the plasmids of the Roseobacter clade.Appl Microbiol Biotechnol. 2013 Apr;97(7):2805-15. doi: 10.1007/s00253-013-4746-8. Epub 2013 Feb 24. Appl Microbiol Biotechnol. 2013. PMID: 23435940 Review.
-
[Research progresses of methanotrophs and methane monooxygenases].Sheng Wu Gong Cheng Xue Bao. 2008 Sep;24(9):1511-9. Sheng Wu Gong Cheng Xue Bao. 2008. PMID: 19160830 Review. Chinese.
Cited by
-
The role of methanotrophy in the microbial carbon metabolism of temperate lakes.Nat Commun. 2022 Jan 10;13(1):43. doi: 10.1038/s41467-021-27718-2. Nat Commun. 2022. PMID: 35013226 Free PMC article.
-
Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (RubisCO) Is Essential for Growth of the Methanotroph Methylococcus capsulatus Strain Bath.Appl Environ Microbiol. 2021 Aug 26;87(18):e0088121. doi: 10.1128/AEM.00881-21. Epub 2021 Aug 26. Appl Environ Microbiol. 2021. PMID: 34288705 Free PMC article.
-
Bioconversion of methane to lactate by an obligate methanotrophic bacterium.Sci Rep. 2016 Feb 23;6:21585. doi: 10.1038/srep21585. Sci Rep. 2016. PMID: 26902345 Free PMC article.
-
Lanthanide-Dependent Regulation of Methanol Oxidation Systems in Methylobacterium extorquens AM1 and Their Contribution to Methanol Growth.J Bacteriol. 2016 Mar 31;198(8):1250-9. doi: 10.1128/JB.00937-15. Print 2016 Apr. J Bacteriol. 2016. PMID: 26833413 Free PMC article.
-
Cre/lox-Mediated CRISPRi Library Reveals Core Genome of a Type I Methanotroph Methylotuvimicrobium buryatense 5GB1C.Appl Environ Microbiol. 2023 Jan 31;89(1):e0188322. doi: 10.1128/aem.01883-22. Epub 2023 Jan 9. Appl Environ Microbiol. 2023. PMID: 36622175 Free PMC article.
References
-
- Shindell D, Kuylenstierna JC, Vignati E, van Dingenen R, Amann M, Klimont Z, Anenberg SC, Muller N, Janssens-Maenhout G, Raes F, Schwartz J, Faluvegi G, Pozzoli L, Kupiainen K, Hoglund-Isaksson L, Emberson L, Streets D, Ramanathan V, Hicks K, Oanh NT, Milly G, Williams M, Demkine V, Fowler D. 2012. Simultaneously mitigating near-term climate change and improving human health and food security. Science 335:183–189. doi:10.1126/science.1210026. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

