Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation
- PMID: 25548173
- PMCID: PMC4299213
- DOI: 10.1073/pnas.1413291112
Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation
Erratum in
-
Correction for Godec et al., Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation.Proc Natl Acad Sci U S A. 2015 Sep 1;112(35):E4968. doi: 10.1073/pnas.1514097112. Epub 2015 Aug 6. Proc Natl Acad Sci U S A. 2015. PMID: 26251345 Free PMC article. No abstract available.
Abstract
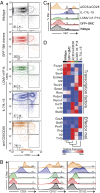
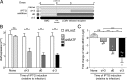
The differentiation of effector CD8(+) T cells is critical for the development of protective responses to pathogens and for effective vaccines. In the first few hours after activation, naive CD8(+) T cells initiate a transcriptional program that leads to the formation of effector and memory T cells, but the regulation of this process is poorly understood. Investigating the role of specific transcription factors (TFs) in determining CD8(+) effector T-cell fate by gene knockdown with RNAi is challenging because naive T cells are refractory to transduction with viral vectors without extensive ex vivo stimulation, which obscures the earliest events in effector differentiation. To overcome this obstacle, we developed a novel strategy to test the function of genes in naive CD8(+) T cells in vivo by creating bone marrow chimera from hematopoietic progenitors transduced with an inducible shRNA construct. Following hematopoietic reconstitution, this approach allowed inducible in vivo gene knockdown in any cell type that developed from this transduced progenitor pool. We demonstrated that lentivirus-transduced progenitor cells could reconstitute normal hematopoiesis and develop into naive CD8(+) T cells that were indistinguishable from wild-type naive T cells. This experimental system enabled induction of efficient gene knockdown in vivo without subsequent manipulation. We applied this strategy to show that the TF BATF is essential for initial commitment of naive CD8(+) T cells to effector development but becomes dispensable by 72h. This approach makes possible the study of gene function in vivo in unperturbed cells of hematopoietic origin that are refractory to viral transduction.
Keywords: BATF; CD8 T cell; RNAi; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. - PubMed
-
- Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annu Rev Immunol. 2003;21:29–70. - PubMed
-
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111(6):837–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

