Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus
- PMID: 25548189
- PMCID: PMC4299237
- DOI: 10.1073/pnas.1422456112
Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus
Abstract
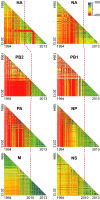
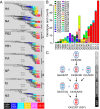
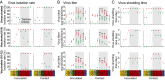
The emergence of human infection with a novel H7N9 influenza virus in China raises a pandemic concern. Chicken H9N2 viruses provided all six of the novel reassortant's internal genes. However, it is not fully understood how the prevalence and evolution of these H9N2 chicken viruses facilitated the genesis of the novel H7N9 viruses. Here we show that over more than 10 y of cocirculation of multiple H9N2 genotypes, a genotype (G57) emerged that had changed antigenicity and improved adaptability in chickens. It became predominant in vaccinated farm chickens in China, caused widespread outbreaks in 2010-2013 before the H7N9 viruses emerged in humans, and finally provided all of their internal genes to the novel H7N9 viruses. The prevalence and variation of H9N2 influenza virus in farmed poultry could provide an important early warning of the emergence of novel reassortants with pandemic potential.
Keywords: H7N9; H9N2; chicken influenza virus; genotype; infectivity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gao R, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–1897. - PubMed
-
- World Health Organization (2014) Human infections with avian influenza A(H7N9) virus. Available at www.who.int/influenza/human_animal_interface/influenza_h7n9/140225_H7N9R.... Accessed December 9, 2014.
-
- Liu D, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: Phylogenetic, structural, and coalescent analyses. Lancet. 2013;381(9881):1926–1932. - PubMed
-
- Wu A, et al. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe. 2013;14(4):446–452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

