A cyclooxygenase-2-dependent prostaglandin E2 biosynthetic system in the Golgi apparatus
- PMID: 25548276
- PMCID: PMC4342474
- DOI: 10.1074/jbc.M114.632463
A cyclooxygenase-2-dependent prostaglandin E2 biosynthetic system in the Golgi apparatus
Abstract
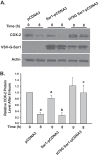
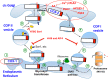
Cyclooxygenases (COXs) catalyze the committed step in prostaglandin (PG) biosynthesis. COX-1 is constitutively expressed and stable, whereas COX-2 is inducible and short lived. COX-2 is degraded via endoplasmic reticulum (ER)-associated degradation (ERAD) following post-translational glycosylation of Asn-594. COX-1 and COX-2 are found in abundance on the luminal surfaces of the ER and inner membrane of the nuclear envelope. Using confocal immunocytofluorescence, we detected both COX-2 and microsomal PGE synthase-1 (mPGES-1) but not COX-1 in the Golgi apparatus. Inhibition of trafficking between the ER and Golgi retarded COX-2 ERAD. COX-2 has a C-terminal STEL sequence, which is an inefficient ER retention signal. Substituting this sequence with KDEL, a robust ER retention signal, concentrated COX-2 in the ER where it was stable and slowly glycosylated on Asn-594. Native COX-2 and a recombinant COX-2 having a Golgi targeting signal but not native COX-1 exhibited efficient catalytic coupling to mPGES-1. We conclude that N-glycosylation of Asn-594 of COX-2 occurs in the ER, leading to anterograde movement of COX-2 to the Golgi where the Asn-594-linked glycan is trimmed prior to retrograde COX-2 transport to the ER for ERAD. Having an inefficient ER retention signal leads to sluggish Golgi to ER transit of COX-2. This permits significant Golgi residence time during which COX-2 can function catalytically. Cytosolic phospholipase A2α, which mobilizes arachidonic acid for PG synthesis, preferentially translocates to the Golgi in response to physiologic Ca(2+) mobilization. We propose that cytosolic phospholipase A2α, COX-2, and mPGES-1 in the Golgi comprise a dedicated system for COX-2-dependent PGE2 biosynthesis.
Keywords: Arachidonic Acid (AA) (ARA); Aspirin; COPII; Coxib; Cyclooxygenase (COX); Degron; Endoplasmic Reticulum-associated Protein Degradation (ERAD); Glycoprotein; NSAID; Prostaglandin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Narumiya S., Furuyashiki T. (2011) Fever, inflammation, pain and beyond: prostanoid receptor research during these 25 years. FASEB J. 25, 813–818 - PubMed
-
- Patrono C., García Rodríguez L. A., Landolfi R., Baigent C. (2005) Low-dose aspirin for the prevention of atherothrombosis. N. Engl. J. Med. 353, 2373–2383 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

