Assessment of difference in gene expression profile between embryos of different derivations
- PMID: 25549061
- PMCID: PMC4312879
- DOI: 10.1089/cell.2014.0057
Assessment of difference in gene expression profile between embryos of different derivations
Abstract
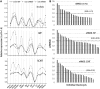
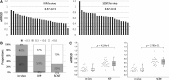
Researchers have exerted sustained efforts to improve the viability of somatic cell nuclear transfer (SCNT) embryos, testing their experimental designs and probing the resultant embryos. However, the lack of a reliable method to estimate the efficacy of these experimental attempts is a chief hindrance to tackling the low-viability problem in SCNT. Here, we introduce a procedure that assesses the degree of difference in gene expression profiles (GEPs) of blastocysts from each other as a representative control of good quality. We first adapted a multiplex reverse transcription-polymerase chain reaction strategy to obtain GEPs for 15 reprogramming-related genes from single mouse blastocysts. GEPs of individual blastocysts displayed a broad range of variations, the extent of which was calculated using a weighted root mean square deviation (wRMSD). wRMSD-based quantitation of GEP difference (qGEP) found that GEP difference between in vivo-derived blastocysts (in vivo) and SCNT blastocysts was greater than the difference between in vivo blastocysts and in vitro-produced (IVP) blastocysts, demonstrating that the SCNT group was more distantly related to the in vivo group than the IVP group. Our qGEP approach for grading individual blastocysts would be useful for selecting a better protocol to derive embryos of better quality prior to field applications.
Figures


References
-
- Amano T., Tani T., Kato Y., and Tsunoda Y. (2001). Mouse cloned from embryonic stem (ES) cells synchronized in metaphase with nocodazole. J. Exp. Zool. 289, 139–145 - PubMed
-
- Amarnath D., Li X., Kato Y., and Tsunoda Y. (2007). Gene expression in individual bovine somatic cell cloned embryos at the 8-cell and blastocyst stages of preimplantation development. J. Reprod. Dev. 53, 1247–1263 - PubMed
-
- Cho S., Park J.S., Kwon S., and Kang Y.K. (2012). Dynamics of Setdb1 expression in early mouse development. Gene Expr. Patterns 12, 213–218 - PubMed
-
- Cho S., Park J.S., and Kang Y.K. (2013). Regulated nuclear entry of over-expressed Setdb1. Genes Cells 18, 694–703 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

