Proteomic screening for amyloid proteins
- PMID: 25549323
- PMCID: PMC4280166
- DOI: 10.1371/journal.pone.0116003
Proteomic screening for amyloid proteins
Abstract
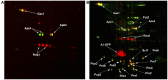
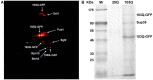
Despite extensive study, progress in elucidation of biological functions of amyloids and their role in pathology is largely restrained due to the lack of universal and reliable biochemical methods for their discovery. All biochemical methods developed so far allowed only identification of glutamine/asparagine-rich amyloid-forming proteins or proteins comprising amyloids that form large deposits. In this article we present a proteomic approach which may enable identification of a broad range of amyloid-forming proteins independently of specific features of their sequences or levels of expression. This approach is based on the isolation of protein fractions enriched with amyloid aggregates via sedimentation by ultracentrifugation in the presence of strong ionic detergents, such as sarkosyl or SDS. Sedimented proteins are then separated either by 2D difference gel electrophoresis or by SDS-PAGE, if they are insoluble in the buffer used for 2D difference gel electrophoresis, after which they are identified by mass-spectrometry. We validated this approach by detection of known yeast prions and mammalian proteins with established capacity for amyloid formation and also revealed yeast proteins forming detergent-insoluble aggregates in the presence of human huntingtin with expanded polyglutamine domain. Notably, with one exception, all these proteins contained glutamine/asparagine-rich stretches suggesting that their aggregates arose due to polymerization cross-seeding by human huntingtin. Importantly, though the approach was developed in a yeast model, it can easily be applied to any organism thus representing an efficient and universal tool for screening for amyloid proteins.
Conflict of interest statement
Figures


References
-
- Sipe JD, Benson MD, Buxbaum JN, Ikeda S, Merlini G, et al. (2012) Amyloid fibril protein nomenclature: 2012 recommendations from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid 19:167–70. - PubMed
-
- Bhak G, Choe Y, Paik S (2009) Mechanism of amyloidogenesis: nucleation-dependent fibrillation versus double-concerted fibrillation. BMB Reports 42:541–51. - PubMed
-
- Prusiner SB (1982) Novel proteinaceous infections particles cause scrapie. Science 216:136–44. - PubMed
-
- Wickner RB (1994) [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae . Science 264:566–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

