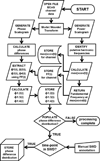
SWDreader: a wavelet-based algorithm using spectral phase to characterize spike-wave morphological variation in genetic models of absence epilepsy
- PMID: 25549550
- PMCID: PMC4331221
- DOI: 10.1016/j.jneumeth.2014.12.016
SWDreader: a wavelet-based algorithm using spectral phase to characterize spike-wave morphological variation in genetic models of absence epilepsy
Abstract
Background: Spike-wave discharges (SWD) found in neuroelectrical recordings are pathognomonic to absence epilepsy. The characteristic spike-wave morphology of the spike-wave complex (SWC) constituents of SWDs can be mathematically described by a subset of possible spectral power and phase values. Morlet wavelet transform (MWT) generates time-frequency representations well-suited to identifying this SWC-associated subset.
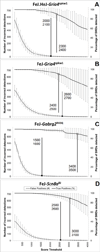
New method: MWT decompositions of SWDs reveal spectral power concentrated at harmonic frequencies. The phase relationships underlying SWC morphology were identified by calculating the differences between phase values at SWD fundamental frequency from the 2nd, 3rd, and 4th harmonics, then using the three phase differences as coordinates to generate a density distribution in a {360°×360°×360°} phase difference space. Strain-specific density distributions were generated from SWDs of mice carrying the Gria4, Gabrg2, or Scn8a mutations to determine whether SWC morphological variants reliably mapped to the same regions of the distribution, and if distribution values could be used to detect SWD.
Comparison with existing methods: To the best of our knowledge, this algorithm is the first to employ spectral phase to quantify SWC morphology, making it possible to computationally distinguish SWC morphological subtypes and detect SWDs.
Results/conclusions: Proof-of-concept testing of the SWDfinder algorithm shows: (1) a major pattern of variation in SWC morphology maps to one axis of the phase difference distribution, (2) variability between the strain-specific distributions reflects differences in the proportions of SWC subtypes generated during SWD, and (3) regularities in the spectral power and phase profiles of SWCs can be used to detect waveforms possessing SWC-like morphology.
Keywords: Absence epilepsy; Fundamental frequency; Harmonic analysis; Morlet wavelet transform; Morphology; Mouse mutant; Phase differences; Seizure detection algorithm; Spike-wave complex; Spike-wave discharge.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures





References
-
- Akman O, Demiralp T, Ates N, Onat FY. Electroencephalographic differences between WAG/Rij and GAERS rat models of absence epilepsy. Epilepsy Research. 2010;89(2–3):185–193. - PubMed
-
- Arneodo A, Audit B, Decoster N, Muzy JF, Vaillant C. The Science of Disasters: Climate Disruptions, Heart Attacks, and Market Crashes. Berlin: Springer; 2002. Wavelet based multifractal formalism: Applications to DNA sequences, satellite images of the cloud structure, and stock market data; pp. 27–96.
-
- Arneodo A, Vaillant C, Audit B, Argoul F, d’Aubenton-Carafa Y, Thermes C. Multi-scale coding of genomic information: From DNA sequence to genome structure and function. Physics Reports. 2011;498(2–3):45–188.
-
- Audit B, Bacry E, Muzy J-F, Arneodo A. Wavelet-based estimators of scaling behavior. IEEE Transactions on Information Theory. 2002;48(11):2938–2954.
-
- Audit B, Baker A, Chen C-L, Rappailles A, Guilbaud G, Julienne H, Goldar A, D’Aubenton-Carafa Y, Hyrien O, Thermes C, Arneodo A. Multiscale analysis of genome-wide replication timing profiles using a wavelet-based signal-processing algorithm. Nature Protocols. 2013;8(1):98–110. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

