In the right place at the right time: visualizing and understanding mRNA localization
- PMID: 25549890
- PMCID: PMC4484810
- DOI: 10.1038/nrm3918
In the right place at the right time: visualizing and understanding mRNA localization
Erratum in
- Nat Rev Mol Cell Biol. 2015 Aug;16(8):513
Abstract
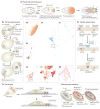
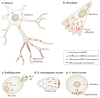
The spatial regulation of protein translation is an efficient way to create functional and structural asymmetries in cells. Recent research has furthered our understanding of how individual cells spatially organize protein synthesis, by applying innovative technology to characterize the relationship between mRNAs and their regulatory proteins, single-mRNA trafficking dynamics, physiological effects of abrogating mRNA localization in vivo and for endogenous mRNA labelling. The implementation of new imaging technologies has yielded valuable information on mRNA localization, for example, by observing single molecules in tissues. The emerging movements and localization patterns of mRNAs in morphologically distinct unicellular organisms and in neurons have illuminated shared and specialized mechanisms of mRNA localization, and this information is complemented by transgenic and biochemical techniques that reveal the biological consequences of mRNA mislocalization.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures



Comment in
-
Translation: Live stream: translation at single-mRNA resolution.Nat Rev Genet. 2016 Jun 16;17(7):373. doi: 10.1038/nrg.2016.79. Nat Rev Genet. 2016. PMID: 27306872 No abstract available.
References
-
- Jeffery WR, Tomlinson CR, Brodeur RD. Localization of actin messenger RNA during early ascidian development. Dev Biol. 1983;99:408–417. - PubMed
-
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. - PubMed
-
- Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopusoocytes. Nature. 1987;328:80–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

