PRSice: Polygenic Risk Score software
- PMID: 25550326
- PMCID: PMC4410663
- DOI: 10.1093/bioinformatics/btu848
PRSice: Polygenic Risk Score software
Abstract
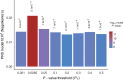
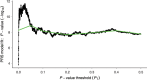
Summary: A polygenic risk score (PRS) is a sum of trait-associated alleles across many genetic loci, typically weighted by effect sizes estimated from a genome-wide association study. The application of PRS has grown in recent years as their utility for detecting shared genetic aetiology among traits has become appreciated; PRS can also be used to establish the presence of a genetic signal in underpowered studies, to infer the genetic architecture of a trait, for screening in clinical trials, and can act as a biomarker for a phenotype. Here we present the first dedicated PRS software, PRSice ('precise'), for calculating, applying, evaluating and plotting the results of PRS. PRSice can calculate PRS at a large number of thresholds ("high resolution") to provide the best-fit PRS, as well as provide results calculated at broad P-value thresholds, can thin Single Nucleotide Polymorphisms (SNPs) according to linkage disequilibrium and P-value or use all SNPs, handles genotyped and imputed data, can calculate and incorporate ancestry-informative variables, and can apply PRS across multiple traits in a single run. We exemplify the use of PRSice via application to data on schizophrenia, major depressive disorder and smoking, illustrate the importance of identifying the best-fit PRS and estimate a P-value significance threshold for high-resolution PRS studies.
Availability and implementation: PRSice is written in R, including wrappers for bash data management scripts and PLINK-1.9 to minimize computational time. PRSice runs as a command-line program with a variety of user-options, and is freely available for download from http://PRSice.info
Contact: jack.euesden@kcl.ac.uk or paul.oreilly@kcl.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

