β-Catenin is central to DUX4-driven network rewiring in facioscapulohumeral muscular dystrophy
- PMID: 25551153
- PMCID: PMC4277075
- DOI: 10.1098/rsif.2014.0797
β-Catenin is central to DUX4-driven network rewiring in facioscapulohumeral muscular dystrophy
Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is an incurable disease, characterized by skeletal muscle weakness and wasting. Genetically, FSHD is characterized by contraction or hypomethylation of repeat D4Z4 units on chromosome 4, which causes aberrant expression of the transcription factor DUX4 from the last repeat. Many genes have been implicated in FSHD pathophysiology, but an integrated molecular model is currently lacking. We developed a novel differential network methodology, Interactome Sparsification and Rewiring (InSpiRe), which detects network rewiring between phenotypes by integrating gene expression data with known protein interactions. Using InSpiRe, we performed a meta-analysis of multiple microarray datasets from FSHD muscle biopsies, then removed secondary rewiring using non-FSHD datasets, to construct a unified network of rewired interactions. Our analysis identified β-catenin as the main coordinator of FSHD-associated protein interaction signalling, with pathways including canonical Wnt, HIF1-α and TNF-α clearly perturbed. To detect transcriptional changes directly elicited by DUX4, gene expression profiling was performed using microarrays on murine myoblasts. This revealed that DUX4 significantly modified expression of the genes in our FSHD network. Furthermore, we experimentally confirmed that Wnt/β-catenin signalling is affected by DUX4 in murine myoblasts. Thus, we provide the first unified molecular map of FSHD signalling, capable of uncovering pathomechanisms and guiding therapeutic development.
Figures

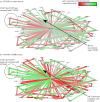
 is the p-value of the statistical analysis performed to determine whether correlation in gene expression between connected edges is different between FSHD and control. Large nodes belong to the core set of 164 high confidence FSHD-specific rewiring genes. Large circles indicate proteins significantly rewiring between FSHD and control phenotypes, identified in the second stage of InSpiRe. There is a clear shift from predominantly uncorrelated to highly correlated between FSHD and controls, with an increased correlation between CTNNB1 and its interaction partners across FSHD samples as compared with controls. This is indicative of increased β-catenin activity. Note the increased positive correlation between CTNNB1 and HIF1A, CASP3 and CASP8.
is the p-value of the statistical analysis performed to determine whether correlation in gene expression between connected edges is different between FSHD and control. Large nodes belong to the core set of 164 high confidence FSHD-specific rewiring genes. Large circles indicate proteins significantly rewiring between FSHD and control phenotypes, identified in the second stage of InSpiRe. There is a clear shift from predominantly uncorrelated to highly correlated between FSHD and controls, with an increased correlation between CTNNB1 and its interaction partners across FSHD samples as compared with controls. This is indicative of increased β-catenin activity. Note the increased positive correlation between CTNNB1 and HIF1A, CASP3 and CASP8.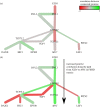


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

