Molecular diagnostic tools for detection and differentiation of phytoplasmas based on chaperonin-60 reveal differences in host plant infection patterns
- PMID: 25551224
- PMCID: PMC4281212
- DOI: 10.1371/journal.pone.0116039
Molecular diagnostic tools for detection and differentiation of phytoplasmas based on chaperonin-60 reveal differences in host plant infection patterns
Abstract
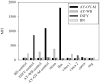
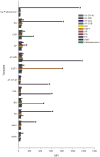
Phytoplasmas ('Candidatus Phytoplasma' spp.) are insect-vectored bacteria that infect a wide variety of plants, including many agriculturally important species. The infections can cause devastating yield losses by inducing morphological changes that dramatically alter inflorescence development. Detection of phytoplasma infection typically utilizes sequences located within the 16S-23S rRNA-encoding locus, and these sequences are necessary for strain identification by currently accepted standards for phytoplasma classification. However, these methods can generate PCR products >1400 bp that are less divergent in sequence than protein-encoding genes, limiting strain resolution in certain cases. We describe a method for accessing the chaperonin-60 (cpn60) gene sequence from a diverse array of 'Ca.Phytoplasma' spp. Two degenerate primer sets were designed based on the known sequence diversity of cpn60 from 'Ca.Phytoplasma' spp. and used to amplify cpn60 gene fragments from various reference samples and infected plant tissues. Forty three cpn60 sequences were thereby determined. The cpn60 PCR-gel electrophoresis method was highly sensitive compared to 16S-23S-targeted PCR-gel electrophoresis. The topology of a phylogenetic tree generated using cpn60 sequences was congruent with that reported for 16S rRNA-encoding genes. The cpn60 sequences were used to design a hybridization array using oligonucleotide-coupled fluorescent microspheres, providing rapid diagnosis and typing of phytoplasma infections. The oligonucleotide-coupled fluorescent microsphere assay revealed samples that were infected simultaneously with two subtypes of phytoplasma. These tools were applied to show that two host plants, Brassica napus and Camelina sativa, displayed different phytoplasma infection patterns.
Conflict of interest statement
Figures


References
-
- Weintraub PG, Beanland L (2006) Insect vectors of phytoplasmas. Annu Rev Entomol. pp.91–111. - PubMed
-
- Gasparich GE (2010) Spiroplasmas and phytoplasmas: microbes associated with plant hosts. Biologicals 38:193–203. - PubMed
-
- Namba S (2011) Phytoplasmas: A century of pioneering research. J Gen Plant Pathol 77:345–349.
-
- Contaldo N, Bertaccini A, Paltrinieri S, Windsor HM, David Windsor G (2012) Axenic culture of plant pathogenic phytoplasmas. Phytopathol Mediterr 51:607–617.
-
- Zhao Y, Davis RE, Wei W, Shao J, Jomantiene R (2014) Phytoplasma Genomes: Evolution Through Mutually Complementary Mechanisms, Gene Loss and Horizontal Acquisition. In: Gross DCeditor. Genomics of Plant-Associated Bacteria. Berlin Heidelberg: Springer-Verlag. pp. 235–271.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

