Antagonistic roles in fetal development and adult physiology for the oppositely imprinted Grb10 and Dlk1 genes
- PMID: 25551289
- PMCID: PMC4280702
- DOI: 10.1186/s12915-014-0099-8
Antagonistic roles in fetal development and adult physiology for the oppositely imprinted Grb10 and Dlk1 genes
Abstract
Background: Despite being a fundamental biological problem the control of body size and proportions during development remains poorly understood, although it is accepted that the insulin-like growth factor (IGF) pathway has a central role in growth regulation, probably in all animals. The involvement of imprinted genes has also attracted much attention, not least because two of the earliest discovered were shown to be oppositely imprinted and antagonistic in their regulation of growth. The Igf2 gene encodes a paternally expressed ligand that promotes growth, while maternally expressed Igf2r encodes a cell surface receptor that restricts growth by sequestering Igf2 and targeting it for lysosomal degradation. There are now over 150 imprinted genes known in mammals, but no other clear examples of antagonistic gene pairs have been identified. The delta-like 1 gene (Dlk1) encodes a putative ligand that promotes fetal growth and in adults restricts adipose deposition. Conversely, Grb10 encodes an intracellular signalling adaptor protein that, when expressed from the maternal allele, acts to restrict fetal growth and is permissive for adipose deposition in adulthood.
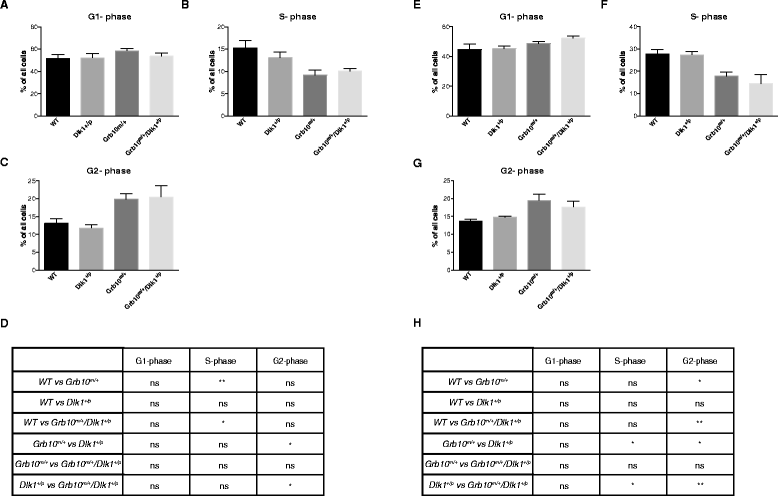
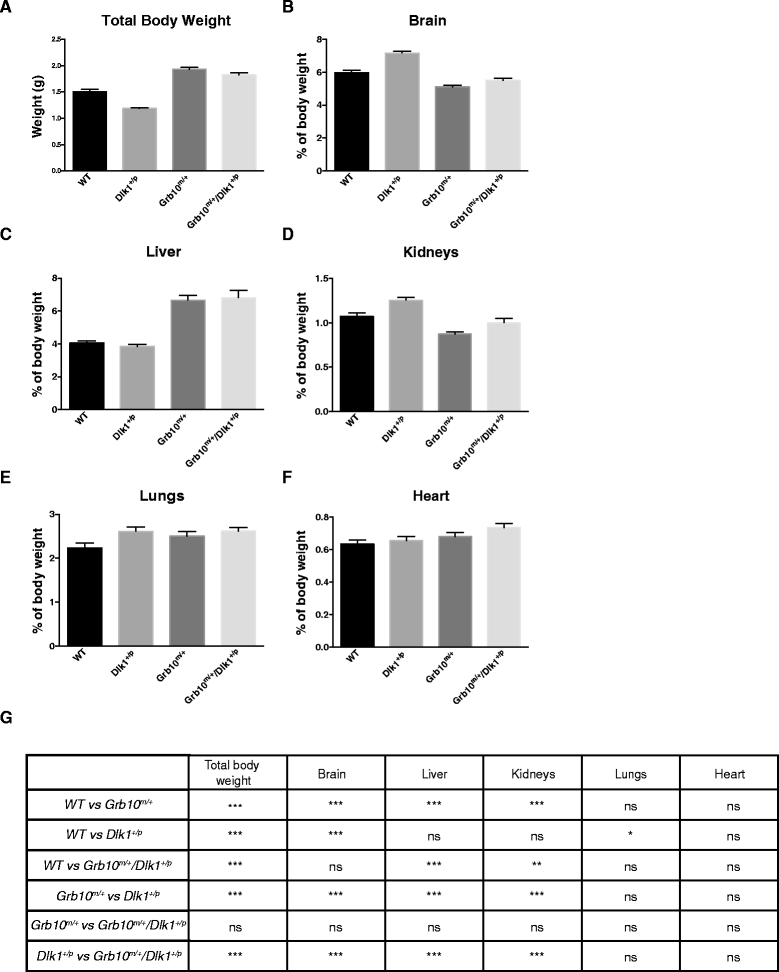
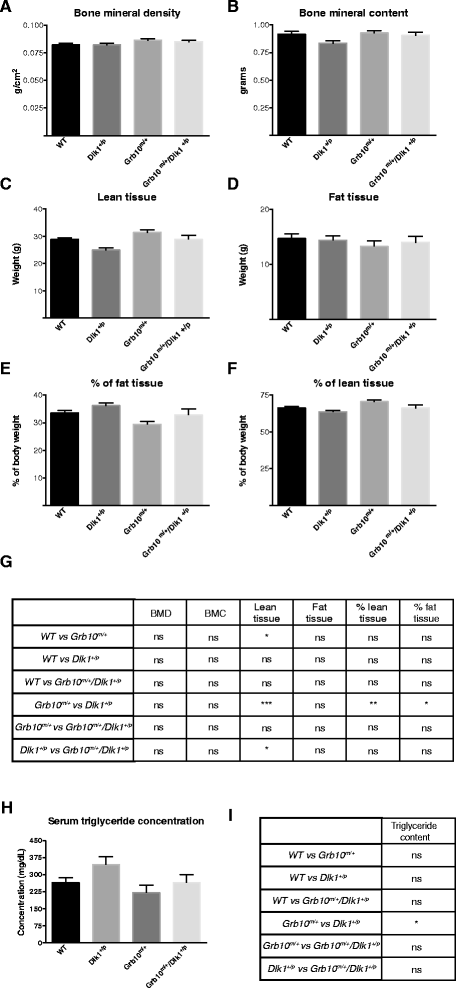
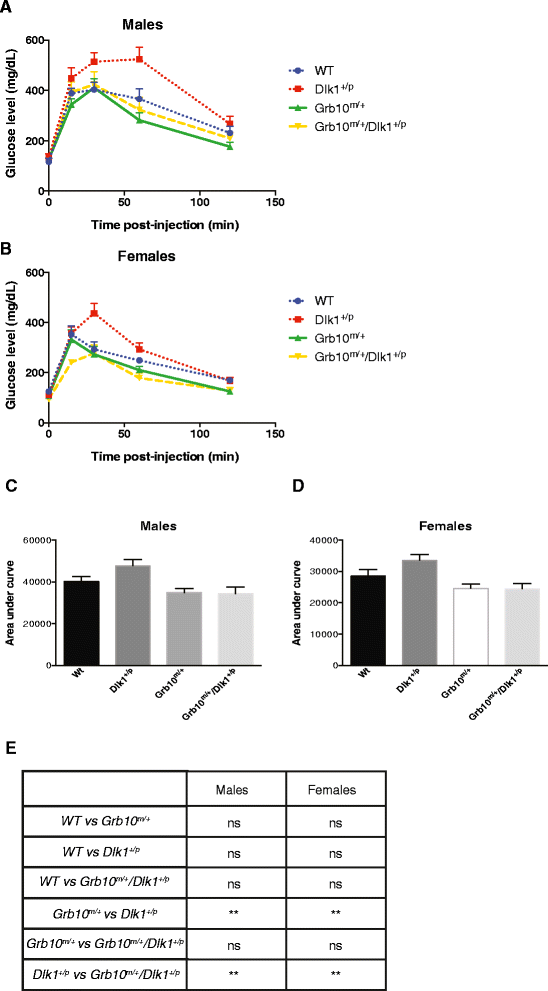
Results: Here, using knockout mice, we present genetic and physiological evidence that these two factors exert their opposite effects on growth and physiology through a common signalling pathway. The major effects are on body size (particularly growth during early life), lean:adipose proportions, glucose regulated metabolism and lipid storage in the liver. A biochemical pathway linking the two cell signalling factors remains to be defined.
Conclusions: We propose that Dlk1 and Grb10 define a mammalian growth axis that is separate from the IGF pathway, yet also features an antagonistic imprinted gene pair.
Figures






Comment in
-
Frugal fat or munificent muscle: genomic imprinting and metabolism.BMC Biol. 2014 Dec 31;12:772. doi: 10.1186/s12915-014-0104-2. BMC Biol. 2014. PMID: 25551395 Free PMC article.
Similar articles
-
Frugal fat or munificent muscle: genomic imprinting and metabolism.BMC Biol. 2014 Dec 31;12:772. doi: 10.1186/s12915-014-0104-2. BMC Biol. 2014. PMID: 25551395 Free PMC article.
-
Imprinted Grb10, encoding growth factor receptor bound protein 10, regulates fetal growth independently of the insulin-like growth factor type 1 receptor (Igf1r) and insulin receptor (Insr) genes.BMC Biol. 2024 May 30;22(1):127. doi: 10.1186/s12915-024-01926-w. BMC Biol. 2024. PMID: 38816743 Free PMC article.
-
The role and interaction of imprinted genes in human fetal growth.Philos Trans R Soc Lond B Biol Sci. 2015 Mar 5;370(1663):20140074. doi: 10.1098/rstb.2014.0074. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25602077 Free PMC article. Review.
-
Grb7, Grb10 and Grb14, encoding the growth factor receptor-bound 7 family of signalling adaptor proteins have overlapping functions in the regulation of fetal growth and post-natal glucose metabolism.BMC Biol. 2024 Sep 30;22(1):221. doi: 10.1186/s12915-024-02018-5. BMC Biol. 2024. PMID: 39343875 Free PMC article.
-
Regulation of growth and metabolism by imprinted genes.Cytogenet Genome Res. 2006;113(1-4):279-91. doi: 10.1159/000090843. Cytogenet Genome Res. 2006. PMID: 16575191 Review.
Cited by
-
Paternal dietary ratio of n-6: n-3 polyunsaturated fatty acids programs offspring leptin expression and gene imprinting in mice.Front Nutr. 2022 Dec 23;9:1043876. doi: 10.3389/fnut.2022.1043876. eCollection 2022. Front Nutr. 2022. PMID: 36618698 Free PMC article.
-
Imprinted Gene Expression and Function of the Dopa Decarboxylase Gene in the Developing Heart.Front Cell Dev Biol. 2021 Jun 22;9:676543. doi: 10.3389/fcell.2021.676543. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34239874 Free PMC article.
-
Dietary protein supplementation and its consequences for intake, digestion, and physical activity of a carnivorous marsupial, Sminthopsis crassicaudata.Ecol Evol. 2018 Mar 5;8(7):3636-3647. doi: 10.1002/ece3.3843. eCollection 2018 Apr. Ecol Evol. 2018. PMID: 29686845 Free PMC article.
-
Frugal fat or munificent muscle: genomic imprinting and metabolism.BMC Biol. 2014 Dec 31;12:772. doi: 10.1186/s12915-014-0104-2. BMC Biol. 2014. PMID: 25551395 Free PMC article.
-
Impulsive Choice in Mice Lacking Paternal Expression of Grb10 Suggests Intragenomic Conflict in Behavior.Genetics. 2018 May;209(1):233-239. doi: 10.1534/genetics.118.300898. Epub 2018 Mar 21. Genetics. 2018. PMID: 29563147 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

