Recognition of the small regulatory RNA RydC by the bacterial Hfq protein
- PMID: 25551292
- PMCID: PMC4337610
- DOI: 10.7554/eLife.05375
Recognition of the small regulatory RNA RydC by the bacterial Hfq protein
Abstract
Bacterial small RNAs (sRNAs) are key elements of regulatory networks that modulate gene expression. The sRNA RydC of Salmonella sp. and Escherichia coli is an example of this class of riboregulators. Like many other sRNAs, RydC bears a 'seed' region that recognises specific transcripts through base-pairing, and its activities are facilitated by the RNA chaperone Hfq. The crystal structure of RydC in complex with E. coli Hfq at a 3.48 Å resolution illuminates how the protein interacts with and presents the sRNA for target recognition. Consolidating the protein-RNA complex is a host of distributed interactions mediated by the natively unstructured termini of Hfq. Based on the structure and other data, we propose a model for a dynamic effector complex comprising Hfq, small RNA, and the cognate mRNA target.
Keywords: E. coli; Hfq; RNA–protein interactions; RydC; biophysics; gene regulation; natively unstructured protein; sRNA; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




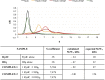


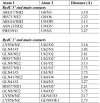
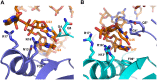



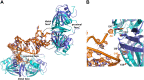

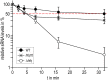



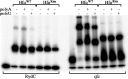
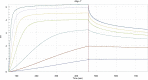
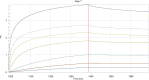



Similar articles
-
Acidic Residues in the Hfq Chaperone Increase the Selectivity of sRNA Binding and Annealing.J Mol Biol. 2015 Nov 6;427(22):3491-3500. doi: 10.1016/j.jmb.2015.07.010. Epub 2015 Jul 18. J Mol Biol. 2015. PMID: 26196441 Free PMC article.
-
Curli synthesis and biofilm formation in enteric bacteria are controlled by a dynamic small RNA module made up of a pseudoknot assisted by an RNA chaperone.Nucleic Acids Res. 2014 Apr;42(7):4682-96. doi: 10.1093/nar/gku098. Epub 2014 Feb 1. Nucleic Acids Res. 2014. PMID: 24489123 Free PMC article.
-
Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq.Biochemistry. 2015 Feb 10;54(5):1157-70. doi: 10.1021/bi500741d. Epub 2015 Jan 26. Biochemistry. 2015. PMID: 25582129
-
Bacterial Small Regulatory RNAs and Hfq Protein.Biochemistry (Mosc). 2015 Dec;80(13):1647-54. doi: 10.1134/S0006297915130027. Biochemistry (Mosc). 2015. PMID: 26878571 Review.
-
Identifying and characterizing Hfq-RNA interactions.Methods. 2013 Sep 15;63(2):144-59. doi: 10.1016/j.ymeth.2013.04.023. Epub 2013 May 23. Methods. 2013. PMID: 23707622 Free PMC article. Review.
Cited by
-
Acidic C-terminal domains autoregulate the RNA chaperone Hfq.Elife. 2017 Aug 9;6:e27049. doi: 10.7554/eLife.27049. Elife. 2017. PMID: 28826489 Free PMC article.
-
Transcriptome-scale analysis uncovers conserved residues in the hydrophobic core of the bacterial RNA chaperone Hfq required for small regulatory RNA stability.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf019. doi: 10.1093/nar/gkaf019. Nucleic Acids Res. 2025. PMID: 39868539 Free PMC article.
-
Diversity of bacterial small RNAs drives competitive strategies for a mutual chaperone.Nat Commun. 2022 May 4;13(1):2449. doi: 10.1038/s41467-022-30211-z. Nat Commun. 2022. PMID: 35508531 Free PMC article.
-
The Bacterial Amyloid-Like Hfq Promotes In Vitro DNA Alignment.Microorganisms. 2019 Dec 3;7(12):639. doi: 10.3390/microorganisms7120639. Microorganisms. 2019. PMID: 31816864 Free PMC article.
-
The binding of Class II sRNA MgrR to two different sites on matchmaker protein Hfq enables efficient competition for Hfq and annealing to regulated mRNAs.RNA. 2018 Dec;24(12):1761-1784. doi: 10.1261/rna.067777.118. Epub 2018 Sep 14. RNA. 2018. PMID: 30217864 Free PMC article.
References
-
- Beich-Frandsen M, Večerek B, Konarev PV, Sjoblom B, Kloiber K, Hammerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, Svergun DI, Konrat R, Bläsi U, Djinovic-Carugo K. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Research. 2011;39:4900–4915. doi: 10.1093/nar/gkq1346. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

