High-affinity DNA-binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling
- PMID: 25552480
- PMCID: PMC4326820
- DOI: 10.1074/jbc.M114.627083
High-affinity DNA-binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling
Abstract
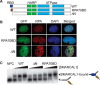
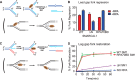
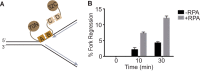
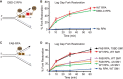
SMARCAL1 catalyzes replication fork remodeling to maintain genome stability. It is recruited to replication forks via an interaction with replication protein A (RPA), the major ssDNA-binding protein in eukaryotic cells. In addition to directing its localization, RPA also activates SMARCAL1 on some fork substrates but inhibits it on others, thereby conferring substrate specificity to SMARCAL1 fork-remodeling reactions. We investigated the mechanism by which RPA regulates SMARCAL1. Our results indicate that although an interaction between SMARCAL1 and RPA is essential for SMARCAL1 activation, the location of the interacting surface on RPA is not. Counterintuitively, high-affinity DNA binding of RPA DNA-binding domain (DBD) A and DBD-B near the fork junction makes it easier for SMARCAL1 to remodel the fork, which requires removing RPA. We also found that RPA DBD-C and DBD-D are not required for SMARCAL1 regulation. Thus, the orientation of the high-affinity RPA DBDs at forks dictates SMARCAL1 substrate specificity.
Keywords: ATPase; DNA Damage Response; DNA Repair; DNA Replication; Fork Regression; Genomic Instability; Replication Protein A; SMARCAL1.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

