Patterns of evolutionary conservation of ascorbic acid-related genes following whole-genome triplication in Brassica rapa
- PMID: 25552535
- PMCID: PMC4316640
- DOI: 10.1093/gbe/evu293
Patterns of evolutionary conservation of ascorbic acid-related genes following whole-genome triplication in Brassica rapa
Abstract
Ascorbic acid (AsA) is an important antioxidant in plants and an essential vitamin for humans. Extending the study of AsA-related genes from Arabidopsis thaliana to Brassica rapa could shed light on the evolution of AsA in plants and inform crop breeding. In this study, we conducted whole-genome annotation, molecular-evolution and gene-expression analyses of all known AsA-related genes in B. rapa. The nucleobase-ascorbate transporter (NAT) gene family and AsA l-galactose pathway genes were also compared among plant species. Four important insights gained are that: 1) 102 AsA-related gene were identified in B. rapa and they mainly diverged 12-18 Ma accompanied by the Brassica-specific genome triplication event; 2) during their evolution, these AsA-related genes were preferentially retained, consistent with the gene dosage hypothesis; 3) the putative proteins were highly conserved, but their expression patterns varied; and 4) although the number of AsA-related genes is higher in B. rapa than in A. thaliana, the AsA contents and the numbers of expressed genes in leaves of both species are similar, the genes that are not generally expressed may serve as substitutes during emergencies. In summary, this study provides genome-wide insights into evolutionary history and mechanisms of AsA-related genes following whole-genome triplication in B. rapa.
Keywords: AsA-related genes; Brassica rapa; evolutionary conservation; expression pattern; gene dosage hypothesis; synteny analysis.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

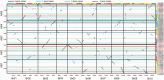
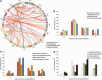


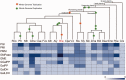



References
-
- Albert VA, et al. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Arrigoni O. Ascorbate system in plant development. J Bioenerg Biomembr. 1994;26:407–419. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

