Population genetics of Anopheles coluzzii immune pathways and genes
- PMID: 25552603
- PMCID: PMC4349087
- DOI: 10.1534/g3.114.014845
Population genetics of Anopheles coluzzii immune pathways and genes
Abstract
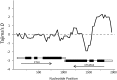
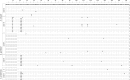
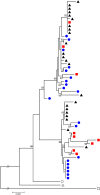
Natural selection is expected to drive adaptive evolution in genes involved in host-pathogen interactions. In this study, we use molecular population genetic analyses to understand how natural selection operates on the immune system of Anopheles coluzzii (formerly A. gambiae "M form"). We analyzed patterns of intraspecific and interspecific genetic variation in 20 immune-related genes and 17 nonimmune genes from a wild population of A. coluzzii and asked if patterns of genetic variation in the immune genes are consistent with pathogen-driven selection shaping the evolution of defense. We found evidence of a balanced polymorphism in CTLMA2, which encodes a C-type lectin involved in regulation of the melanization response. The two CTLMA2 haplotypes, which are distinguished by fixed amino acid differences near the predicted peptide cleavage site, are also segregating in the sister species A. gambiae ("S form") and A. arabiensis. Comparison of the two haplotypes between species indicates that they were not shared among the species through introgression, but rather that they arose before the species divergence and have been adaptively maintained as a balanced polymorphism in all three species. We additionally found that STAT-B, a retroduplicate of STAT-A, shows strong evidence of adaptive evolution that is consistent with neofunctionalization after duplication. In contrast to the striking patterns of adaptive evolution observed in these Anopheles-specific immune genes, we found no evidence of adaptive evolution in the Toll and Imd innate immune pathways that are orthologously conserved throughout insects. Genes encoding the Imd pathway exhibit high rates of amino acid divergence between Anopheles species but also display elevated amino acid diversity that is consistent with relaxed purifying selection. These results indicate that adaptive coevolution between A. coluzzii and its pathogens is more likely to involve novel or lineage-specific molecular mechanisms than the canonical humoral immune pathways.
Keywords: C-type lectin; JAK-STAT; balancing selection; genetics of immunity; innate immunity; population genetics.
Copyright © 2015 Rottschaefer et al.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
