Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies
- PMID: 25552646
- PMCID: PMC4375422
- DOI: 10.1093/hmg/ddu733
Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies
Abstract
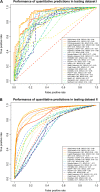
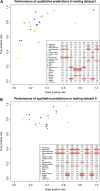
Accurate deleteriousness prediction for nonsynonymous variants is crucial for distinguishing pathogenic mutations from background polymorphisms in whole exome sequencing (WES) studies. Although many deleteriousness prediction methods have been developed, their prediction results are sometimes inconsistent with each other and their relative merits are still unclear in practical applications. To address these issues, we comprehensively evaluated the predictive performance of 18 current deleteriousness-scoring methods, including 11 function prediction scores (PolyPhen-2, SIFT, MutationTaster, Mutation Assessor, FATHMM, LRT, PANTHER, PhD-SNP, SNAP, SNPs&GO and MutPred), 3 conservation scores (GERP++, SiPhy and PhyloP) and 4 ensemble scores (CADD, PON-P, KGGSeq and CONDEL). We found that FATHMM and KGGSeq had the highest discriminative power among independent scores and ensemble scores, respectively. Moreover, to ensure unbiased performance evaluation of these prediction scores, we manually collected three distinct testing datasets, on which no current prediction scores were tuned. In addition, we developed two new ensemble scores that integrate nine independent scores and allele frequency. Our scores achieved the highest discriminative power compared with all the deleteriousness prediction scores tested and showed low false-positive prediction rate for benign yet rare nonsynonymous variants, which demonstrated the value of combining information from multiple orthologous approaches. Finally, to facilitate variant prioritization in WES studies, we have pre-computed our ensemble scores for 87 347 044 possible variants in the whole-exome and made them publicly available through the ANNOVAR software and the dbNSFP database.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Ng P.C., Henikoff S. (2006) Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet., 7, 61–80. - PubMed
-
- Thusberg J., Vihinen M. (2009) Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum. Mutat., 30, 703–714. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

