Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein
- PMID: 25552709
- PMCID: PMC4337552
- DOI: 10.1128/JVI.03125-14
Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein
Abstract
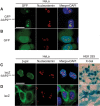
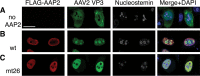
Assembly-activating protein (AAP) of adeno-associated virus serotype 2 (AAV2) is a nucleolar-localizing protein that plays a critical role in transporting the viral capsid VP3 protein to the nucleolus for assembly. Here, we identify and characterize AAV2 AAP (AAP2) nuclear (NLS) and nucleolar (NoLS) localization signals near the carboxy-terminal region of AAP2 (amino acid positions 144 to 184) (AAP2(144-184)). This region contains five basic-amino-acid-rich (BR) clusters, KSKRSRR (AAP2BR1), RRR (AAP2BR2), RFR (AAP2BR3), RSTSSR (AAP2BR4), and RRIK (AAP2BR5), from the amino terminus to the carboxy terminus. We created 30 AAP2BR mutants by arginine/lysine-to-alanine mutagenesis or deletion of AAP2BRs and 8 and 1 green fluorescent protein (GFP)-AAP2BR and β-galactosidase-AAP2BR fusion proteins, respectively, and analyzed their intracellular localization in HeLa cells by immunofluorescence microscopy. The results showed that AAP2(144-184) has redundant multipartite NLSs and that any combinations of 4 AAP2BRs, but not 3 or less, can constitute a functional NLS-NoLS; AAP2BR1 and AAP2BR2 play the most influential role for nuclear localization, but either one of the two AAP2BRs is dispensable if all 4 of the other AAP2BRs are present, resulting in 3 different, overlapping NLS motifs; and the NoLS is shared redundantly among the five AAP2BRs and functions in a context-dependent manner. AAP2BR mutations not only resulted in aberrant intracellular localization, but also attenuated AAP2 protein expression to various degrees, and both of these abnormalities have a significant negative impact on capsid production. Thus, this study reveals the organization of the intermingling NLSs and NoLSs in AAP2 and provides insights into their functional roles in capsid assembly.
Importance: Adeno-associated virus (AAV) has become a popular and successful vector for in vivo gene therapy; however, its biology has yet to be fully understood. In this regard, the recent discovery of the assembly-activating protein (AAP), a nonstructural, nucleolar-localizing AAV protein essential for viral capsid assembly, has provided us a new opportunity to better understand the fundamental processes required for virion formation. Here, we identify clusters of basic amino acids in the carboxy terminus of AAP from AAV serotype 2 (AAV2) that act as nuclear and nucleolar localization signals. We also demonstrate their importance in maintaining AAP expression levels and efficient production of viral capsids. Insights into the functions of AAP can elucidate the requirements and process for AAV capsid assembly, which may lead to improved vector production for use in gene therapy. This study also contributes to the growing body of work on nuclear and nucleolar localization signals.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
Adeno-associated Virus (AAV) Assembly-Activating Protein Is Not an Essential Requirement for Capsid Assembly of AAV Serotypes 4, 5, and 11.J Virol. 2017 Jan 18;91(3):e01980-16. doi: 10.1128/JVI.01980-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27852862 Free PMC article.
-
Mapping and Engineering Functional Domains of the Assembly-Activating Protein of Adeno-associated Viruses.J Virol. 2018 Jun 29;92(14):e00393-18. doi: 10.1128/JVI.00393-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29695425 Free PMC article.
-
Nuclear translocation of adeno-associated virus type 2 capsid proteins for virion assembly.J Gen Virol. 2012 Sep;93(Pt 9):1887-1898. doi: 10.1099/vir.0.043232-0. Epub 2012 Jun 13. J Gen Virol. 2012. PMID: 22694902
-
The role of nuclear localization signal in parvovirus life cycle.Virol J. 2017 Apr 14;14(1):80. doi: 10.1186/s12985-017-0745-1. Virol J. 2017. PMID: 28410597 Free PMC article. Review.
-
Structural and cellular biology of adeno-associated virus attachment and entry.Adv Virus Res. 2020;106:39-84. doi: 10.1016/bs.aivir.2020.01.002. Epub 2020 Feb 13. Adv Virus Res. 2020. PMID: 32327148 Review.
Cited by
-
Functional roles of the membrane-associated AAV protein MAAP.Sci Rep. 2021 Nov 4;11(1):21698. doi: 10.1038/s41598-021-01220-7. Sci Rep. 2021. PMID: 34737404 Free PMC article.
-
A Quantitative Dot Blot Assay for AAV Titration and Its Use for Functional Assessment of the Adeno-associated Virus Assembly-activating Proteins.J Vis Exp. 2018 Jun 12;(136):56766. doi: 10.3791/56766. J Vis Exp. 2018. PMID: 29985340 Free PMC article.
-
The Assembly-Activating Protein Promotes Stability and Interactions between AAV's Viral Proteins to Nucleate Capsid Assembly.Cell Rep. 2018 May 8;23(6):1817-1830. doi: 10.1016/j.celrep.2018.04.026. Cell Rep. 2018. PMID: 29742436 Free PMC article.
-
Assembly of Structurally Simple Icosahedral Viruses.Subcell Biochem. 2024;105:403-430. doi: 10.1007/978-3-031-65187-8_11. Subcell Biochem. 2024. PMID: 39738953 Review.
-
The membrane associated accessory protein is an adeno-associated viral egress factor.Nat Commun. 2021 Oct 29;12(1):6239. doi: 10.1038/s41467-021-26485-4. Nat Commun. 2021. PMID: 34716331 Free PMC article.
References
-
- Sonntag F, Kother K, Schmidt K, Weghofer M, Raupp C, Nieto K, Kuck A, Gerlach B, Bottcher B, Muller OJ, Lux K, Horer M, Kleinschmidt JA. 2011. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol 85:12686–12697. doi:10.1128/JVI.05359-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

