RNA-mediated epigenetic regulation of gene expression
- PMID: 25554358
- PMCID: PMC4376354
- DOI: 10.1038/nrg3863
RNA-mediated epigenetic regulation of gene expression
Abstract
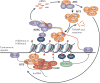
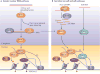
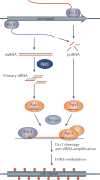
Diverse classes of RNA, ranging from small to long non-coding RNAs, have emerged as key regulators of gene expression, genome stability and defence against foreign genetic elements. Small RNAs modify chromatin structure and silence transcription by guiding Argonaute-containing complexes to complementary nascent RNA scaffolds and then mediating the recruitment of histone and DNA methyltransferases. In addition, recent advances suggest that chromatin-associated long non-coding RNA scaffolds also recruit chromatin-modifying complexes independently of small RNAs. These co-transcriptional silencing mechanisms form powerful RNA surveillance systems that detect and silence inappropriate transcription events, and provide a memory of these events via self-reinforcing epigenetic loops.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Cech TR, Steitz JA. The noncoding RNA revolution — trashing old rules to forge new ones. Cell. 2014;157:77–94. - PubMed
-
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. - PubMed
-
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. - PubMed
-
- Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by mi RNAs and siRNAs. Mol Cell. 2004;15:185–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

