Scaling of gene expression with transcription-factor fugacity
- PMID: 25554908
- PMCID: PMC4386862
- DOI: 10.1103/PhysRevLett.113.258101
Scaling of gene expression with transcription-factor fugacity
Abstract
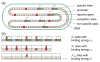
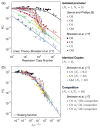
The proteins associated with gene regulation are often shared between multiple pathways simultaneously. By way of contrast, models in regulatory biology often assume these pathways act independently. We demonstrate a framework for calculating the change in gene expression for the interacting case by decoupling repressor occupancy across the cell from the gene of interest by way of a chemical potential. The details of the interacting regulatory architecture are encompassed in an effective concentration, and thus, a single scaling function describes a collection of gene expression data from diverse regulatory situations and collapses it onto a single master curve.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
