Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle
- PMID: 25555795
- PMCID: PMC4331006
- DOI: 10.15252/embj.201488307
Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle
Abstract
Eukaryotic replication origins are defined by the ORC-dependent loading of the Mcm2-7 helicase complex onto chromatin in G1. Paradoxically, there is a vast excess of Mcm2-7 relative to ORC assembled onto chromatin in G1. These excess Mcm2-7 complexes exhibit little co-localization with ORC or replication foci and can function as dormant origins. We dissected the mechanisms regulating the assembly and distribution of the Mcm2-7 complex in the Drosophila genome. We found that in the absence of cyclin E/Cdk2 activity, there was a 10-fold decrease in chromatin-associated Mcm2-7 relative to the levels found at the G1/S transition. The minimal amounts of Mcm2-7 loaded in the absence of cyclin E/Cdk2 activity were strictly localized to ORC binding sites. In contrast, cyclin E/Cdk2 activity was required for maximal loading of Mcm2-7 and a dramatic genome-wide reorganization of the distribution of Mcm2-7 that is shaped by active transcription. Thus, increasing cyclin E/Cdk2 activity over the course of G1 is not only critical for Mcm2-7 loading, but also for the distribution of the Mcm2-7 helicase prior to S-phase entry.
Keywords: DNA replication; Mcm2‐7; cell cycle; chromatin.
© 2015 The Authors.
Figures

FACS profiles of DNA content for cells arrested at different points in the cell cycle by Dup/Cdt1 RNAi, cyclin E RNAi, Cdk2 RNAi, Dacapo overexpression (+Dacapo), and 1 mM HU.
Analysis of Mcm2-7 nuclear chromatin association at different points in the cell cycle. Nuclear chromatin fractions and cytoplasmic extracts were assayed for Orc2 and Mcm2-7 by Western blot. A non-specific band is indicated by an asterisk (*).
Quantification of the ratio of chromatin-bound Mcm2-7 relative to Orc2 (log10 scale) for a minimum of 11 replicates (mean ± SD).
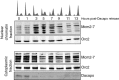

Schematic of the experiment.
FACS profiles of DNA content for each cell population assayed.
Western blot analysis for Orc2 and Mcm2-7 in the nuclear chromatin fraction and Orc2, Mcm2-7, Dup/Cdt1, and cyclin E in the cytoplasmic fraction for each condition assayed.
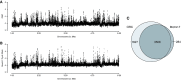
Genome-wide analysis of ORC localization by ChIP-chip. ORC enrichment from asynchronous cells is depicted for a 5-Mb section of chromosome 2L.
Genome-wide analysis of Mcm2-7 localization in early G1 by ChIP-chip. Mcm2-7 enrichment from cyclin E RNAi-depleted cells is depicted for a 5-Mb section of chromosome 2L.
Venn diagram depicting the overlap between ORC and Mcm2-7 peaks.
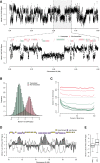
Genome-wide analysis of Mcm2-7 localization at the G1/S transition by ChIP-chip. Mcm2-7 enrichment from HU-arrested cells is depicted for a 5-Mb section of chromosome 2L. Inset: transcribed (green) and non-transcribed (red) genes are indicated above with genes on the positive strand on the top and those on the negative on the bottom.
Bimodal distribution of Mcm2-7 enrichment over transcribed and non-transcribed genes. Histogram showing the distribution of probe scores found within transcribed (green) and non-transcribed (red) genes.
“Meta”-gene analysis of Mcm2-7 enrichment for different deciles of gene expression and their aggregated probe intensities.
Mcm2-7 is displaced from chromatin by DNA replication. Genome-wide analysis of Mcm2-7 localization in late S-phase (6 h post HU release) by ChIP-chip. Mcm2-7 enrichment is depicted for a 10-Mb section of chromosome 2L (filled gray), replication timing profile (black line) and early (yellow) and late (purple) replication timing domains.
Box-plots representing late S-phase Mcm2-7 ChIP signal found within early (246) or late (167) replication domains.
References
-
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. - PubMed
-
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. - PubMed
-
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

