An N-terminal extension to the hepatitis B virus core protein forms a poorly ordered trimeric spike in assembled virus-like particles
- PMID: 25557498
- PMCID: PMC4318616
- DOI: 10.1016/j.jsb.2014.12.006
An N-terminal extension to the hepatitis B virus core protein forms a poorly ordered trimeric spike in assembled virus-like particles
Abstract
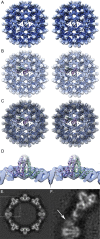
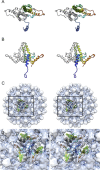
Virus-like particles composed of the core antigen of hepatitis B virus (HBcAg) have been shown to be an effective platform for the display of foreign epitopes in vaccine development. Heterologous sequences have been successfully inserted at both amino and carboxy termini as well as internally at the major immunodominant epitope. We used cryogenic electron microscopy (CryoEM) and three-dimensional image reconstruction to investigate the structure of VLPs assembled from an N-terminal extended HBcAg that contained a polyhistidine tag. The insert was seen to form a trimeric spike on the capsid surface that was poorly resolved, most likely owing to it being flexible. We hypothesise that the capacity of N-terminal inserts to form trimers may have application in the development of multivalent vaccines to trimeric antigens. Our analysis also highlights the value of tools for local resolution assessment in studies of partially disordered macromolecular assemblies by cryoEM.
Keywords: Cryo-electron microscopy; Hepatitis B virus; Local resolution; Three-dimensional reconstruction; Vaccine; Virus-like particle.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
3.5Å cryoEM structure of hepatitis B virus core assembled from full-length core protein.PLoS One. 2013 Sep 6;8(9):e69729. doi: 10.1371/journal.pone.0069729. eCollection 2013. PLoS One. 2013. PMID: 24039702 Free PMC article.
-
Enhanced stability of a chimeric hepatitis B core antigen virus-like-particle (HBcAg-VLP) by a C-terminal linker-hexahistidine-peptide.J Nanobiotechnology. 2018 Apr 13;16(1):39. doi: 10.1186/s12951-018-0363-0. J Nanobiotechnology. 2018. PMID: 29653575 Free PMC article.
-
Structural basis for the development of avian virus capsids that display influenza virus proteins and induce protective immunity.J Virol. 2015 Mar;89(5):2563-74. doi: 10.1128/JVI.03025-14. Epub 2014 Dec 17. J Virol. 2015. PMID: 25520499 Free PMC article.
-
Hybrid hepatitis B virus core antigen as a vaccine carrier moiety: I. presentation of foreign epitopes.J Biotechnol. 1996 Jan 26;44(1-3):91-6. doi: 10.1016/0168-1656(95)00118-2. J Biotechnol. 1996. PMID: 8717391 Review.
-
New chimaeric hepatitis B virus core particles carrying hantavirus (serotype Puumala) epitopes: immunogenicity and protection against virus challenge.J Biotechnol. 1999 Aug 20;73(2-3):141-53. doi: 10.1016/s0168-1656(99)00117-0. J Biotechnol. 1999. PMID: 10486924 Review.
Cited by
-
Multifaceted virus-like particles: Navigating towards broadly effective influenza A virus vaccines.Curr Res Microb Sci. 2024 Nov 15;8:100317. doi: 10.1016/j.crmicr.2024.100317. eCollection 2025. Curr Res Microb Sci. 2024. PMID: 39717209 Free PMC article. Review.
-
Epitope selection and their placement for increased virus neutralization in a novel vaccination strategy for porcine epidemic diarrhea virus utilizing the Hepatitis B virus core antigen.Vaccine. 2018 Jul 16;36(30):4507-4516. doi: 10.1016/j.vaccine.2018.06.015. Epub 2018 Jun 15. Vaccine. 2018. PMID: 29914846 Free PMC article.
-
Hepatitis B virus Core protein nuclear interactome identifies SRSF10 as a host RNA-binding protein restricting HBV RNA production.PLoS Pathog. 2020 Nov 12;16(11):e1008593. doi: 10.1371/journal.ppat.1008593. eCollection 2020 Nov. PLoS Pathog. 2020. PMID: 33180834 Free PMC article.
-
Needle-free, spirulina-produced Plasmodium falciparum circumsporozoite vaccination provides sterile protection against pre-erythrocytic malaria in mice.NPJ Vaccines. 2022 Oct 4;7(1):113. doi: 10.1038/s41541-022-00534-5. NPJ Vaccines. 2022. PMID: 36195607 Free PMC article.
-
A Simple Add-and-Display Method for Immobilisation of Cancer Drug on His-tagged Virus-like Nanoparticles for Controlled Drug Delivery.Sci Rep. 2017 Jul 13;7(1):5303. doi: 10.1038/s41598-017-05525-4. Sci Rep. 2017. PMID: 28706267 Free PMC article.
References
-
- Adrian M., Dubochet J., Lepault J., McDowall A.W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. - PubMed
-
- Baker T.S., Cheng R.H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. - PubMed
-
- Crowther R.A., Amos L.A., Finch J.T., De Rosier D.J., Klug A. Three dimensional reconstructions of spherical viruses by fourier synthesis from electron micrographs. Nature. 1970;226:421–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

