Biased allelic expression in human primary fibroblast single cells
- PMID: 25557783
- PMCID: PMC4289680
- DOI: 10.1016/j.ajhg.2014.12.001
Biased allelic expression in human primary fibroblast single cells
Abstract
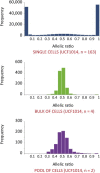
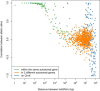
The study of gene expression in mammalian single cells via genomic technologies now provides the possibility to investigate the patterns of allelic gene expression. We used single-cell RNA sequencing to detect the allele-specific mRNA level in 203 single human primary fibroblasts over 133,633 unique heterozygous single-nucleotide variants (hetSNVs). We observed that at the snapshot of analyses, each cell contained mostly transcripts from one allele from the majority of genes; indeed, 76.4% of the hetSNVs displayed stochastic monoallelic expression in single cells. Remarkably, adjacent hetSNVs exhibited a haplotype-consistent allelic ratio; in contrast, distant sites located in two different genes were independent of the haplotype structure. Moreover, the allele-specific expression in single cells correlated with the abundance of the cellular transcript. We observed that genes expressing both alleles in the majority of the single cells at a given time point were rare and enriched with highly expressed genes. The relative abundance of each allele in a cell was controlled by some regulatory mechanisms given that we observed related single-cell allelic profiles according to genes. Overall, these results have direct implications in cellular phenotypic variability.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Ferguson-Smith A.C. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011;12:565–575. - PubMed
-
- Reik W., Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. - PubMed
-
- Augui S., Nora E.P., Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. - PubMed
-
- Chow J.C., Yen Z., Ziesche S.M., Brown C.J. Silencing of the mammalian X chromosome. Annu. Rev. Genomics Hum. Genet. 2005;6:69–92. - PubMed
-
- Chess A., Simon I., Cedar H., Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

