Developing discriminate model and comparative analysis of differentially expressed genes and pathways for bloodstream samples of diabetes mellitus type 2
- PMID: 25559614
- PMCID: PMC4304197
- DOI: 10.1186/1471-2105-15-S17-S5
Developing discriminate model and comparative analysis of differentially expressed genes and pathways for bloodstream samples of diabetes mellitus type 2
Abstract
Background: Diabetes mellitus of type 2 (T2D), also known as noninsulin-dependent diabetes mellitus (NIDDM) or adult-onset diabetes, is a common disease. It is estimated that more than 300 million people worldwide suffer from T2D. In this study, we investigated the T2D, pre-diabetic and healthy human (no diabetes) bloodstream samples using genomic, genealogical, and phonemic information. We identified differentially expressed genes and pathways. The study has provided deeper insights into the development of T2D, and provided useful information for further effective prevention and treatment of the disease.
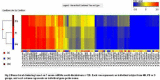
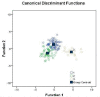
Results: A total of 142 bloodstream samples were collected, including 47 healthy humans, 22 pre-diabetic and 73 T2D patients. Whole genome scale gene expression profiles were obtained using the Agilent Oligo chips that contain over 20,000 human genes. We identified 79 significantly differentially expressed genes that have fold change ≥ 2. We mapped those genes and pinpointed locations of those genes on human chromosomes. Amongst them, 3 genes were not mapped well on the human genome, but the rest of 76 differentially expressed genes were well mapped on the human genome. We found that most abundant differentially expressed genes are on chromosome one, which contains 9 of those genes, followed by chromosome two that contains 7 of the 76 differentially expressed genes. We performed gene ontology (GO) functional analysis of those 79 differentially expressed genes and found that genes involve in the regulation of cell proliferation were among most common pathways related to T2D. The expression of the 79 genes was combined with clinical information that includes age, sex, and race to construct an optimal discriminant model. The overall performance of the model reached 95.1% accuracy, with 91.5% accuracy on identifying healthy humans, 100% accuracy on pre-diabetic patients and 95.9% accuract on T2D patients. The higher performance on identifying pre-diabetic patients was resulted from more significant changes of gene expressions among this particular group of humans, which implicated that patients were having profound genetic changes towards disease development.
Conclusion: Differentially expressed genes were distributed across chromosomes, and are more abundant on chromosomes 1 and 2 than the rest of the human genome. We found that regulation of cell proliferation actually plays an important role in the T2D disease development. The predictive model developed in this study has utilized the 79 significant genes in combination with age, sex, and racial information to distinguish pre-diabetic, T2D, and healthy humans. The study not only has provided deeper understanding of the disease molecular mechanisms but also useful information for pathway analysis and effective drug target identification.
Figures

Similar articles
-
Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes.Acta Diabetol. 2016 Oct;53(5):693-702. doi: 10.1007/s00592-016-0837-1. Epub 2016 Apr 2. Acta Diabetol. 2016. PMID: 27039347
-
Identification of genes and pathways involved in kidney renal clear cell carcinoma.BMC Bioinformatics. 2014;15 Suppl 17(Suppl 17):S2. doi: 10.1186/1471-2105-15-S17-S2. Epub 2014 Dec 16. BMC Bioinformatics. 2014. PMID: 25559354 Free PMC article.
-
Gene expression profiles displayed by peripheral blood mononuclear cells from patients with type 2 diabetes mellitus focusing on biological processes implicated on the pathogenesis of the disease.Gene. 2012 Dec 15;511(2):151-60. doi: 10.1016/j.gene.2012.09.090. Epub 2012 Oct 2. Gene. 2012. PMID: 23036710
-
Genetics of type II diabetes.Recent Prog Horm Res. 1998;53:201-16. Recent Prog Horm Res. 1998. PMID: 9769709 Review.
-
Shared genetic etiology underlying Alzheimer's disease and type 2 diabetes.Mol Aspects Med. 2015 Jun-Oct;43-44:66-76. doi: 10.1016/j.mam.2015.06.006. Epub 2015 Jun 23. Mol Aspects Med. 2015. PMID: 26116273 Free PMC article. Review.
Cited by
-
Clinical relevance of epigenetics in the onset and management of type 2 diabetes mellitus.Epigenetics. 2017 Jun 3;12(6):401-415. doi: 10.1080/15592294.2016.1278097. Epub 2017 Jan 6. Epigenetics. 2017. PMID: 28059593 Free PMC article. Review.
-
Advances in translational bioinformatics facilitate revealing the landscape of complex disease mechanisms.BMC Bioinformatics. 2014;15 Suppl 17(Suppl 17):I1. doi: 10.1186/1471-2105-15-S17-I1. Epub 2014 Dec 16. BMC Bioinformatics. 2014. PMID: 25559210 Free PMC article.
References
-
- World Health Organization. http://www.who.int/diabetes/facts/world_figures/en/index.html
-
- Centers for Disease Control and Prevention. http://www.cdc.gov/diabetes/pubs/pdf/ndfs_20
-
- From the Centers for Disease Control and Prevention. http://www.cdc.gov/mmwr/preview/M
-
- Letchuman GR, Wan NW, Wan MW, Chandran LR, Tee GH, Jamaiyah H, Isa MR, Zanariah H, Fatanah I, Ahmad FY. Prevalence of diabetes in the Malaysian National Health Morbidity Survey III 2006. Med J Malaysia. 2010;65:180–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

