Steatosis-induced proteins adducts with lipid peroxidation products and nuclear electrophilic stress in hepatocytes
- PMID: 25560244
- PMCID: PMC4309854
- DOI: 10.1016/j.redox.2014.12.009
Steatosis-induced proteins adducts with lipid peroxidation products and nuclear electrophilic stress in hepatocytes
Abstract
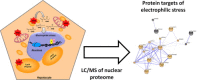
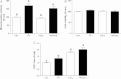
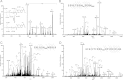
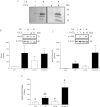
Accumulating evidence suggests that fatty livers are particularly more susceptible to several pathological conditions, including hepatic inflammation, cirrhosis and liver cancer. However the exact mechanism of such susceptibility is still largely obscure. The current study aimed to elucidate the effect of hepatocytes lipid accumulation on the nuclear electrophilic stress. Accumulation of intracellular lipids was significantly increased in HepG2 cells incubated with fatty acid (FA) complex (1mM, 2:1 oleic and palmitic acids). In FA-treated cells, lipid droplets were localized around the nucleus and seemed to induce mechanical force, leading to the disruption of the nucleus morphology. Level of reactive oxygen species (ROS) was significantly increased in FA-loaded cells and was further augmented by treatment with moderate stressor (CoCl2). Increased ROS resulted in formation of reactive carbonyls (aldehydes and ketones, derived from lipid peroxidation) with a strong perinuclear accumulation. Mass-spectroscopy analysis indicated that lipid accumulation per-se can results in modification of nuclear protein by reactive lipid peroxidation products (oxoLPP). 235 Modified proteins involved in transcription regulation, splicing, protein synthesis and degradation, DNA repair and lipid metabolism were identified uniquely in FA-treated cells. These findings suggest that steatosis can affect nuclear redox state, and induce modifications of nuclear proteins by reactive oxoLPP accumulated in the perinuclear space upon FA-treatment.
Keywords: Electrophilic stress; Fatty liver diseases; Lipid peroxidation; Protein modifications; Reactive carbonyls.
Copyright © 2014 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

