Insights into the origin and evolution of the plant hormone signaling machinery
- PMID: 25560880
- PMCID: PMC4348752
- DOI: 10.1104/pp.114.247403
Insights into the origin and evolution of the plant hormone signaling machinery
Abstract
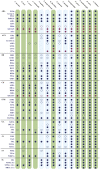
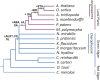
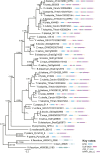
Plant hormones modulate plant growth, development, and defense. However, many aspects of the origin and evolution of plant hormone signaling pathways remain obscure. Here, we use a comparative genomic and phylogenetic approach to investigate the origin and evolution of nine major plant hormone (abscisic acid, auxin, brassinosteroid, cytokinin, ethylene, gibberellin, jasmonate, salicylic acid, and strigolactone) signaling pathways. Our multispecies genome-wide analysis reveals that: (1) auxin, cytokinin, and strigolactone signaling pathways originated in charophyte lineages; (2) abscisic acid, jasmonate, and salicylic acid signaling pathways arose in the last common ancestor of land plants; (3) gibberellin signaling evolved after the divergence of bryophytes from land plants; (4) the canonical brassinosteroid signaling originated before the emergence of angiosperms but likely after the split of gymnosperms and angiosperms; and (5) the origin of the canonical ethylene signaling pathway postdates shortly the emergence of angiosperms. Our findings might have important implications in understanding the molecular mechanisms underlying the emergence of land plants.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. - PubMed
-
- Boursiac Y, Léran S, Corratgé-Faillie C, Gojon A, Krouk G, Lacombe B (2013) ABA transport and transporters. Trends Plant Sci 18: 325–333 - PubMed
-
- Bowman JL. (2013) Walkabout on the long branches of plant evolution. Curr Opin Plant Biol 16: 70–77 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

