A new antibiotic kills pathogens without detectable resistance
- PMID: 25561178
- PMCID: PMC7414797
- DOI: 10.1038/nature14098
A new antibiotic kills pathogens without detectable resistance
Erratum in
-
Erratum: A new antibiotic kills pathogens without detectable resistance.Nature. 2015 Apr 16;520(7547):388. doi: 10.1038/nature14303. Epub 2015 Feb 25. Nature. 2015. PMID: 25731174 No abstract available.
Abstract
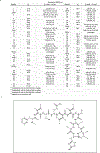
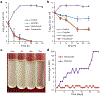
Antibiotic resistance is spreading faster than the introduction of new compounds into clinical practice, causing a public health crisis. Most antibiotics were produced by screening soil microorganisms, but this limited resource of cultivable bacteria was overmined by the 1960s. Synthetic approaches to produce antibiotics have been unable to replace this platform. Uncultured bacteria make up approximately 99% of all species in external environments, and are an untapped source of new antibiotics. We developed several methods to grow uncultured organisms by cultivation in situ or by using specific growth factors. Here we report a new antibiotic that we term teixobactin, discovered in a screen of uncultured bacteria. Teixobactin inhibits cell wall synthesis by binding to a highly conserved motif of lipid II (precursor of peptidoglycan) and lipid III (precursor of cell wall teichoic acid). We did not obtain any mutants of Staphylococcus aureus or Mycobacterium tuberculosis resistant to teixobactin. The properties of this compound suggest a path towards developing antibiotics that are likely to avoid development of resistance.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
Figures





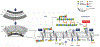




Comment in
-
Antibiotics: An irresistible newcomer.Nature. 2015 Jan 22;517(7535):442-4. doi: 10.1038/nature14193. Epub 2015 Jan 7. Nature. 2015. PMID: 25561172 No abstract available.
-
Antibacterial drugs: a new drug for resistant bugs.Nat Rev Drug Discov. 2015 Feb;14(2):94. doi: 10.1038/nrd4544. Nat Rev Drug Discov. 2015. PMID: 25633792 No abstract available.
-
Drug discovery: Early antibiotic from a cranberry bog.Nature. 2015 Feb 19;518(7539):303. doi: 10.1038/518303a. Nature. 2015. PMID: 25693555 No abstract available.
-
Bacteria: Assessing resistance to new antibiotics.Nature. 2015 Mar 12;519(7542):158. doi: 10.1038/519158e. Nature. 2015. PMID: 25762274 No abstract available.
References
-
- Fleming A On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol 10, 226–236 (1929).
-
- Kardos N & Demain AL Penicillin: the medicine with the greatest impact on therapeutic outcomes. Appl. Microbiol. Biotechnol 92, 677–687 (2011). - PubMed
-
- Schatz A, Bugie E & Waksman SA Streptomycin, a substance exhibiting antibiotic activity againstgram-positive and gram-negative bacteria. Proc. Soc. Exp. Biol. Med 55, 66–69 (1944). - PubMed
-
- Spellberg B & Shlaes D Prioritized current unmet needs for antibacterial therapies. Clin. Pharmacol. Ther 96, 151–153 (2014). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

