Improvement of the fluorescence intensity during a flow cytometric analysis for rice protoplasts by localization of a green fluorescent protein into chloroplasts
- PMID: 25561231
- PMCID: PMC4307275
- DOI: 10.3390/ijms16010788
Improvement of the fluorescence intensity during a flow cytometric analysis for rice protoplasts by localization of a green fluorescent protein into chloroplasts
Abstract
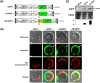
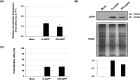
Protoplasts have been a useful unicellular system for various molecular biological analyses based on transient expression and single cell analysis using fluorescence-activated cell sorting (FACS), widely used as a powerful method in functional genomics. Despite the versatility of these methods, some limits based on low fluorescence intensity of a flow cytometric analysis (FCA) using protoplasts have been reported. In this study, the chloroplast targeting of fluorescent proteins (FPs) led to an eight-fold increase in fluorescence intensity and a 4.5-fold increase of transfection ratio from 14.7% to 65.7% as compared with their targeting into the cytoplasm. Moreover, the plot data of FCA shows that 83.3% of the K-sGFP population is under the threshold level, regarded as a non-transgenic population with background signals, while 65.7% of the K-sGFP population is spread on overall intervals. To investigate the reason underlying this finding, mRNA/protein levels and transfection efficiency were analyzed, and results suggest that mRNA/protein levels and transfection ratio are not much different between K-sGFP and KR-sGFP. From those results, we hypothesized that the difference of fluorescence intensity is not only derived from cellular events such as molecular level or transfection efficiency. Taken together, we suggest that the translocation of FPs into chloroplasts contributes to the improvement of fluorescence intensity in FCA and, apparently, plays an important role in minimizing the loss of the transfected population. Our study could be usefully applicable for highly sensitive FACS and FCA-investigations of green tissue.
Figures




Similar articles
-
Plastoglobule-Targeting Competence of a Putative Transit Peptide Sequence from Rice Phytoene Synthase 2 in Plastids.Int J Mol Sci. 2016 Dec 22;18(1):18. doi: 10.3390/ijms18010018. Int J Mol Sci. 2016. PMID: 28025520 Free PMC article.
-
Transient expression and analysis of chloroplast proteins in Arabidopsis protoplasts.Methods Mol Biol. 2011;774:59-71. doi: 10.1007/978-1-61779-234-2_4. Methods Mol Biol. 2011. PMID: 21822832
-
[Efficient transient expression to analyze miRNA targets in rice protoplasts].Sheng Wu Gong Cheng Xue Bao. 2014 Nov;30(11):1751-62. Sheng Wu Gong Cheng Xue Bao. 2014. PMID: 25985526 Chinese.
-
Multiparametric analysis, sorting, and transcriptional profiling of plant protoplasts and nuclei according to cell type.Methods Mol Biol. 2011;699:407-29. doi: 10.1007/978-1-61737-950-5_20. Methods Mol Biol. 2011. PMID: 21116995 Review.
-
Rice peptide deformylase PDF1B is crucial for development of chloroplasts.Plant Cell Physiol. 2008 Oct;49(10):1536-46. doi: 10.1093/pcp/pcn121. Epub 2008 Aug 20. Plant Cell Physiol. 2008. PMID: 18718933
Cited by
-
Chlorophyll Fluorescence Video Imaging: A Versatile Tool for Identifying Factors Related to Photosynthesis.Front Plant Sci. 2018 Jan 30;9:55. doi: 10.3389/fpls.2018.00055. eCollection 2018. Front Plant Sci. 2018. PMID: 29472935 Free PMC article. Review.
-
A Versatile High Throughput Screening Platform for Plant Metabolic Engineering Highlights the Major Role of ABI3 in Lipid Metabolism Regulation.Front Plant Sci. 2020 Mar 17;11:288. doi: 10.3389/fpls.2020.00288. eCollection 2020. Front Plant Sci. 2020. PMID: 32256511 Free PMC article.
-
Genome-Wide Analysis of Cyclic Nucleotide-Gated Channel Genes Related to Pollen Development in Rice.Plants (Basel). 2022 Nov 17;11(22):3145. doi: 10.3390/plants11223145. Plants (Basel). 2022. PMID: 36432876 Free PMC article.
-
OsYSL13 Is Involved in Iron Distribution in Rice.Int J Mol Sci. 2018 Nov 9;19(11):3537. doi: 10.3390/ijms19113537. Int J Mol Sci. 2018. PMID: 30423990 Free PMC article.
-
Enhanced Expression of QTL qLL9/DEP1 Facilitates the Improvement of Leaf Morphology and Grain Yield in Rice.Int J Mol Sci. 2019 Feb 17;20(4):866. doi: 10.3390/ijms20040866. Int J Mol Sci. 2019. PMID: 30781568 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

