Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana
- PMID: 25561503
- PMCID: PMC4349977
- DOI: 10.1074/mcp.M114.040352
Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana
Abstract
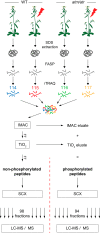
The reversible phosphorylation of proteins on serine, threonine, and tyrosine residues is an important biological regulatory mechanism. In the context of genome integrity, signaling cascades driven by phosphorylation are crucial for the coordination and regulation of DNA repair. The two serine/threonine protein kinases ataxia telangiectasia-mutated (ATM) and Ataxia telangiectasia-mutated and Rad3-related (ATR) are key factors in this process, each specific for different kinds of DNA lesions. They are conserved across eukaryotes, mediating the activation of cell-cycle checkpoints, chromatin modifications, and regulation of DNA repair proteins. We designed a novel mass spectrometry-based phosphoproteomics approach to study DNA damage repair in Arabidopsis thaliana. The protocol combines filter aided sample preparation, immobilized metal affinity chromatography, metal oxide affinity chromatography, and strong cation exchange chromatography for phosphopeptide generation, enrichment, and separation. Isobaric labeling employing iTRAQ (isobaric tags for relative and absolute quantitation) was used for profiling the phosphoproteome of atm atr double mutants and wild type plants under either regular growth conditions or challenged by irradiation. A total of 10,831 proteins were identified and 15,445 unique phosphopeptides were quantified, containing 134 up- and 38 down-regulated ATM/ATR dependent phosphopeptides. We identified known and novel ATM/ATR targets such as LIG4 and MRE11 (needed for resistance against ionizing radiation), PIE1 and SDG26 (implicated in chromatin remodeling), PCNA1, WAPL, and PDS5 (implicated in DNA replication), and ASK1 and HTA10 (involved in meiosis).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related.Mol Biol Cell. 2005 May;16(5):2566-76. doi: 10.1091/mbc.e04-10-0890. Epub 2005 Mar 16. Mol Biol Cell. 2005. PMID: 15772150 Free PMC article.
-
SOG1 transcription factor promotes the onset of endoreduplication under salinity stress in Arabidopsis.Sci Rep. 2021 Jun 2;11(1):11659. doi: 10.1038/s41598-021-91293-1. Sci Rep. 2021. Retraction in: Sci Rep. 2024 Feb 21;14(1):4282. doi: 10.1038/s41598-024-54831-1. PMID: 34079040 Free PMC article. Retracted.
-
Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint.Plant Cell. 2007 Jan;19(1):211-25. doi: 10.1105/tpc.106.045047. Epub 2007 Jan 5. Plant Cell. 2007. PMID: 17209125 Free PMC article.
-
An insight into the mechanism of DNA damage response in plants- role of SUPPRESSOR OF GAMMA RESPONSE 1: An overview.Mutat Res. 2020 Jan-Apr;819-820:111689. doi: 10.1016/j.mrfmmm.2020.111689. Epub 2020 Jan 23. Mutat Res. 2020. PMID: 32004947 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
-
Highly ABA-Induced 1 (HAI1)-Interacting protein HIN1 and drought acclimation-enhanced splicing efficiency at intron retention sites.Proc Natl Acad Sci U S A. 2019 Oct 29;116(44):22376-22385. doi: 10.1073/pnas.1906244116. Epub 2019 Oct 14. Proc Natl Acad Sci U S A. 2019. PMID: 31611386 Free PMC article.
-
Salinity stress-induced phosphorylation of INDETERMINATE-DOMAIN 4 (IDD4) by MPK6 regulates plant growth adaptation in Arabidopsis.Front Plant Sci. 2023 Oct 10;14:1265687. doi: 10.3389/fpls.2023.1265687. eCollection 2023. Front Plant Sci. 2023. PMID: 37881611 Free PMC article.
-
Thylakoid Protein Phosphorylation Dynamics in a Moss Mutant Lacking SERINE/THREONINE PROTEIN KINASE STN8.Plant Physiol. 2019 Jul;180(3):1582-1597. doi: 10.1104/pp.19.00117. Epub 2019 May 6. Plant Physiol. 2019. PMID: 31061101 Free PMC article.
-
Analysis of state 1-state 2 transitions by genome editing and complementation reveals a quenching component independent from the formation of PSI-LHCI-LHCII supercomplex in Arabidopsis thaliana.Biol Direct. 2023 Aug 23;18(1):49. doi: 10.1186/s13062-023-00406-5. Biol Direct. 2023. PMID: 37612770 Free PMC article.
-
Long noncoding RNAs contribute to DNA damage resistance in Arabidopsis thaliana.Genetics. 2023 Aug 31;225(1):iyad135. doi: 10.1093/genetics/iyad135. Genetics. 2023. PMID: 37467473 Free PMC article.
References
-
- Dissmeyer N., Schnittger A. (2011) Guide to the book plant kinases. Methods Mol. Biol. 779, 3–5 - PubMed
-
- Greenwell P. W., Kronmal S. L., Porter S. E., Gassenhuber J., Obermaier B., Petes T. D. (1995) TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82, 823–829 - PubMed
-
- Weinert T. A., Kiser G. L., Hartwell L. H. (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8, 652–665 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

