Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry
- PMID: 25561506
- PMCID: PMC4349991
- DOI: 10.1074/mcp.M113.035550
Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry
Abstract
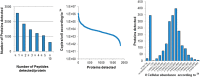
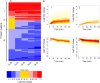
Targeted mass spectrometry by selected reaction monitoring (S/MRM) has proven to be a suitable technique for the consistent and reproducible quantification of proteins across multiple biological samples and a wide dynamic range. This performance profile is an important prerequisite for systems biology and biomedical research. However, the method is limited to the measurements of a few hundred peptides per LC-MS analysis. Recently, we introduced SWATH-MS, a combination of data independent acquisition and targeted data analysis that vastly extends the number of peptides/proteins quantified per sample, while maintaining the favorable performance profile of S/MRM. Here we applied the SWATH-MS technique to quantify changes over time in a large fraction of the proteome expressed in Saccharomyces cerevisiae in response to osmotic stress. We sampled cell cultures in biological triplicates at six time points following the application of osmotic stress and acquired single injection data independent acquisition data sets on a high-resolution 5600 tripleTOF instrument operated in SWATH mode. Proteins were quantified by the targeted extraction and integration of transition signal groups from the SWATH-MS datasets for peptides that are proteotypic for specific yeast proteins. We consistently identified and quantified more than 15,000 peptides and 2500 proteins across the 18 samples. We demonstrate high reproducibility between technical and biological replicates across all time points and protein abundances. In addition, we show that the abundance of hundreds of proteins was significantly regulated upon osmotic shock, and pathway enrichment analysis revealed that the proteins reacting to osmotic shock are mainly involved in the carbohydrate and amino acid metabolism. Overall, this study demonstrates the ability of SWATH-MS to efficiently generate reproducible, consistent, and quantitatively accurate measurements of a large fraction of a proteome across multiple samples.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Addona T. A., Shi X., Keshishian H., Mani D. R., Burgess M., Gillette M. A., Clauser K. R., Shen D., Lewis G. D., Farrell L. A., Fifer M. A., Sabatine M. S., Gerszten R. E., Carr S. A. (2011) A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nat. Biotechnol. 29, 635–643 - PMC - PubMed
-
- Picotti P., Clement-Ziza M., Lam H., Campbell D. S., Schmidt A., Deutsch E. W., Rost H., Sun Z., Rinner O., Reiter L., Shen Q., Michaelson J. J., Frei A., Alberti S., Kusebauch U., Wollscheid B., Moritz R. L., Beyer A., Aebersold R. (2013) A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 494, 266–270 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

