Histone variants and epigenetics
- PMID: 25561719
- PMCID: PMC4292162
- DOI: 10.1101/cshperspect.a019364
Histone variants and epigenetics
Abstract
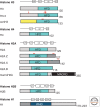
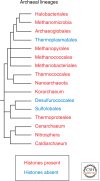
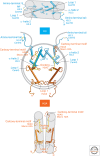
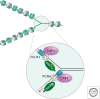
Histones package and compact DNA by assembling into nucleosome core particles. Most histones are synthesized at S phase for rapid deposition behind replication forks. In addition, the replacement of histones deposited during S phase by variants that can be deposited independently of replication provide the most fundamental level of chromatin differentiation. Alternative mechanisms for depositing different variants can potentially establish and maintain epigenetic states. Variants have also evolved crucial roles in chromosome segregation, transcriptional regulation, DNA repair, and other processes. Investigations into the evolution, structure, and metabolism of histone variants provide a foundation for understanding the participation of chromatin in important cellular processes and in epigenetic memory.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
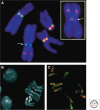
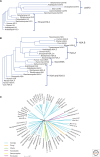
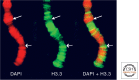



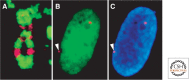
References
-
- Akhtar A, Gasser SM 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–517. - PubMed
WWW RESOURCES
-
- http://www.ebi.ac.uk/Tools/phylogeny. EBI server.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
