Structure of neuroblastoma suppressor of tumorigenicity 1 (NBL1): insights for the functional variability across bone morphogenetic protein (BMP) antagonists
- PMID: 25561725
- PMCID: PMC4335214
- DOI: 10.1074/jbc.M114.628412
Structure of neuroblastoma suppressor of tumorigenicity 1 (NBL1): insights for the functional variability across bone morphogenetic protein (BMP) antagonists
Abstract
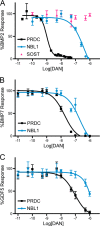
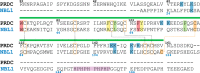
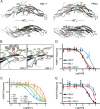
Bone morphogenetic proteins (BMPs) are antagonized through the action of numerous extracellular protein antagonists, including members from the differential screening-selected gene aberrative in neuroblastoma (DAN) family. In vivo, misregulation of the balance between BMP signaling and DAN inhibition can lead to numerous disease states, including cancer, kidney nephropathy, and pulmonary arterial hypertension. Despite this importance, very little information is available describing how DAN family proteins effectively inhibit BMP ligands. Furthermore, our understanding for how differences in individual DAN family members arise, including affinity and specificity, remains underdeveloped. Here, we present the structure of the founding member of the DAN family, neuroblastoma suppressor of tumorigenicity 1 (NBL1). Comparing NBL1 to the structure of protein related to Dan and Cerberus (PRDC), a more potent BMP antagonist within the DAN family, a number of differences were identified. Through a mutagenesis-based approach, we were able to correlate the BMP binding epitope in NBL1 with that in PRDC, where introduction of specific PRDC amino acids in NBL1 (A58F and S67Y) correlated with a gain-of-function inhibition toward BMP2 and BMP7, but not GDF5. Although NBL1(S67Y) was able to antagonize BMP7 as effectively as PRDC, NBL1(S67Y) was still 32-fold weaker than PRDC against BMP2. Taken together, this data suggests that alterations in the BMP binding epitope can partially account for differences in the potency of BMP inhibition within the DAN family.
Keywords: Bone Morphogenetic Protein (BMP); Differential Screening-aberrative in Neuroblastoma (DAN); Extracellular Antagonism; Mutagenesis; Neuroblastoma Suppressor of Tumorigenicity 1 (NBL1); Protein Related to Dan and Cerberus (PRDC); SOST; Structural Biology; Transforming Growth Factor β(TGF-B); X-ray Crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Bragdon B., Moseychuk O., Saldanha S., King D., Julian J., Nohe A. (2011) Bone morphogenetic proteins: a critical review. Cell Signal. 23, 609–620 - PubMed
-
- Cai J., Pardali E., Sánchez-Duffhues G., ten Dijke P. (2012) BMP signaling in vascular diseases. FEBS Lett. 586, 1993–2002 - PubMed
-
- Walsh D. W., Godson C., Brazil D. P., Martin F. (2010) Extracellular BMP-antagonist regulation in development and disease: tied up in knots. Trends Cell Biol. 20, 244–256 - PubMed
-
- Rider C. C., Mulloy B. (2010) Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem. J. 429, 1–12 - PubMed
-
- Tardif G., Pelletier J.-P., Boileau C., Martel-Pelletier J. (2009) The BMP antagonists Follistatin and Gremlin in normal and early osteoarthritic cartilage: an immunohistochemical study. Osteoarthr. Cartil. 17, 263–270 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

