Identification of Stably Expressed lncRNAs as Valid Endogenous Controls for Profiling of Human Glioma
- PMID: 25561975
- PMCID: PMC4280393
- DOI: 10.7150/jca.10867
Identification of Stably Expressed lncRNAs as Valid Endogenous Controls for Profiling of Human Glioma
Abstract
Background: Recent research indicates that long non-coding RNAs (lncRNA) represent a new family of RNAs that is of fundamental importance for controlling transcription and translation. Thereby, there is increasing evidence that lncRNAs are also important in tumourigenesis. Thereby valid expression profiling using quantitative PCR requires suitable, stably expressed normalisers to achieve reliable and reproducible data. However, no systematic analysis of suitable references in lncRNA studies in human glioma has been performed yet.
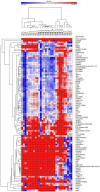
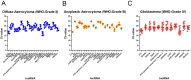
Methods: In this study, we investigated 90 lncRNAs in 30 tissue specimen for the expression stability in human diffuse astrocytoma (WHO-Grade II), anaplastic astrocytoma (WHO-Grade III) and glioblastoma (WHO-Grade IV) both alone as well as in comparison with normal white matter. Our identification procedure included a rigorous bioinformatical selection process that resulted in the inclusion of only highly abundant, equally expressed lncRNAs for further analysis. Additionally, lncRNAs were classified according to their stability value using the NormFinder algorithm.
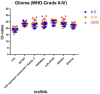
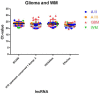
Results: We identified 24 appropriate normalisers suitable for studies in diffuse astrocytoma, 22 for studies in anaplastic astrocytoma and 12 for studies in glioblastoma. Comparing all three glioma entities 7 lncRNAs showed stable expression levels. Addition of normal brain tissue resulted in only 4 suitable lncRNAs.
Conclusions: Our findings indicate that 4 lncRNAs (HOXA6as, H19 upstream conserved 1 and 2, Zfhx2as and BC200) are suitable as normalisers in glioma and normal brain. These lncRNAs may thus be regarded as universal references being applicable for the accurate normalisation of lncRNA expression profiling in various glioma (WHO-Grades II-IV) alone and in combination with brain tissue. This enables to perform valid longitudinal studies, e.g. of glioma before and after malignisation to identify changes of lncRNA expressions probably driving malignant transformation.
Keywords: Glioma; Profiling.; References; lncRNA; long non-coding RNA; qPCR.
Conflict of interest statement
Competing Interests: None declared.
Figures
Similar articles
-
Long non-coding RNA normalisers in human brain tissue.J Neural Transm (Vienna). 2015 Jul;122(7):1045-54. doi: 10.1007/s00702-014-1352-6. Epub 2014 Dec 21. J Neural Transm (Vienna). 2015. PMID: 25528156
-
ACTB and SDHA Are Suitable Endogenous Reference Genes for Gene Expression Studies in Human Astrocytomas Using Quantitative RT-PCR.Technol Cancer Res Treat. 2018 Jan 1;17:1533033818802318. doi: 10.1177/1533033818802318. Technol Cancer Res Treat. 2018. PMID: 30259794 Free PMC article.
-
Identification of valid endogenous control genes for determining gene expression in human glioma.Neuro Oncol. 2010 Jun;12(6):570-9. doi: 10.1093/neuonc/nop072. Epub 2010 Feb 5. Neuro Oncol. 2010. PMID: 20511187 Free PMC article.
-
Expression of lncRNAs in glioma: A lighthouse for patients with glioma.Heliyon. 2024 Jan 24;10(3):e24799. doi: 10.1016/j.heliyon.2024.e24799. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38322836 Free PMC article. Review.
-
Non-Coding RNAs in Glioma.Cancers (Basel). 2018 Dec 22;11(1):17. doi: 10.3390/cancers11010017. Cancers (Basel). 2018. PMID: 30583549 Free PMC article. Review.
Cited by
-
Genetic Characterization of Ten-Eleven-Translocation Methylcytosine Dioxygenase Alterations in Human Glioma.J Cancer. 2015 Jul 15;6(9):832-42. doi: 10.7150/jca.12010. eCollection 2015. J Cancer. 2015. PMID: 26284134 Free PMC article.
-
Long noncoding RNAs and Alzheimer's disease.Clin Interv Aging. 2016 Jun 29;11:867-72. doi: 10.2147/CIA.S107037. eCollection 2016. Clin Interv Aging. 2016. PMID: 27418812 Free PMC article. Review.
-
Altered Long Noncoding RNA Expression Precedes the Course of Parkinson's Disease-a Preliminary Report.Mol Neurobiol. 2017 May;54(4):2869-2877. doi: 10.1007/s12035-016-9854-x. Epub 2016 Mar 28. Mol Neurobiol. 2017. PMID: 27021022
-
Up-Regulation of Long Non-Coding RNA AB073614 Predicts a Poor Prognosis in Patients with Glioma.Int J Environ Res Public Health. 2016 Apr 19;13(4):433. doi: 10.3390/ijerph13040433. Int J Environ Res Public Health. 2016. PMID: 27104549 Free PMC article.
-
Long Non-Coding RNAs in Gliomas: From Molecular Pathology to Diagnostic Biomarkers and Therapeutic Targets.Int J Mol Sci. 2018 Sep 13;19(9):2754. doi: 10.3390/ijms19092754. Int J Mol Sci. 2018. PMID: 30217088 Free PMC article. Review.
References
-
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. BioEssays: news and reviews in molecular, cellular and developmental biology. 2009;31:51–9. doi:10.1002/bies.080099. - PubMed
-
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155–9. doi:10.1038/nrg2521. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

