Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities
- PMID: 25563503
- PMCID: PMC4307746
- DOI: 10.1186/s40246-014-0023-x
Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities
Erratum in
-
Correction to: Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities.Hum Genomics. 2021 Jun 15;15(1):35. doi: 10.1186/s40246-021-00322-7. Hum Genomics. 2021. PMID: 34127067 Free PMC article. No abstract available.
Abstract
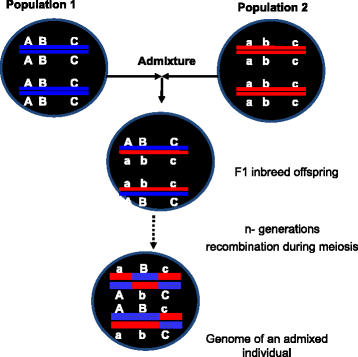
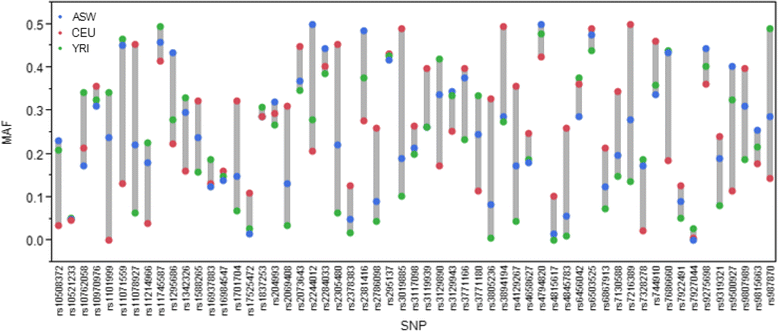
This review explores the limitations of self-reported race, ethnicity, and genetic ancestry in biomedical research. Various terminologies are used to classify human differences in genomic research including race, ethnicity, and ancestry. Although race and ethnicity are related, race refers to a person's physical appearance, such as skin color and eye color. Ethnicity, on the other hand, refers to communality in cultural heritage, language, social practice, traditions, and geopolitical factors. Genetic ancestry inferred using ancestry informative markers (AIMs) is based on genetic/genomic data. Phenotype-based race/ethnicity information and data computed using AIMs often disagree. For example, self-reporting African Americans can have drastically different levels of African or European ancestry. Genetic analysis of individual ancestry shows that some self-identified African Americans have up to 99% of European ancestry, whereas some self-identified European Americans have substantial admixture from African ancestry. Similarly, African ancestry in the Latino population varies between 3% in Mexican Americans to 16% in Puerto Ricans. The implication of this is that, in African American or Latino populations, self-reported ancestry may not be as accurate as direct assessment of individual genomic information in predicting treatment outcomes. To better understand human genetic variation in the context of health disparities, we suggest using "ancestry" (or biogeographical ancestry) to describe actual genetic variation, "race" to describe health disparity in societies characterized by racial categories, and "ethnicity" to describe traditions, lifestyle, diet, and values. We also suggest using ancestry informative markers for precise characterization of individuals' biological ancestry. Understanding the sources of human genetic variation and the causes of health disparities could lead to interventions that would improve the health of all individuals.
Figures




References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

