Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin β1 and PI3K
- PMID: 25563751
- PMCID: PMC5379035
- DOI: 10.1038/srep07656
Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin β1 and PI3K
Abstract
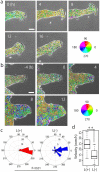
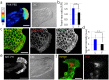
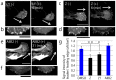
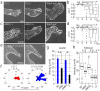
Collective cell migration plays a crucial role in several biological processes, such as embryonic development, wound healing, and cancer metastasis. Here, we focused on collectively migrating Madin-Darby Canine Kidney (MDCK) epithelial cells that follow a leader cell on a collagen gel to clarify the mechanism of collective cell migration. First, we removed a leader cell from the migrating collective with a micromanipulator. This then caused disruption of the cohesive migration of cells that followed in movement, called "follower" cells, which showed the importance of leader cells. Next, we observed localization of active Rac, integrin β1, and PI3K. These molecules were clearly localized in the leading edge of leader cells, but not in follower cells. Live cell imaging using active Rac and active PI3K indicators was performed to elucidate the relationship between Rac, integrin β1, and PI3K. Finally, we demonstrated that the inhibition of these molecules resulted in the disruption of collective migration. Our findings not only demonstrated the significance of a leader cell in collective cell migration, but also showed that Rac, integrin β1, and PI3K are upregulated in leader cells and drive collective cell migration.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Friedl P. & Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10, 445–57 (2009). - PubMed
-
- Friedl P., Hegerfeldt Y. & Tusch M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol 48, 441–9 (2004). - PubMed
-
- Rorth P. Collective cell migration. Annu Rev Cell Dev Biol 25, 407–29 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

