Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome
- PMID: 25563792
- PMCID: PMC4331925
- DOI: 10.1186/1471-2164-15-S12-S4
Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome
Abstract
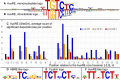
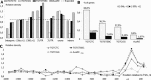
Auxin responsive elements (AuxRE) were found in upstream regions of target genes for ARFs (Auxin response factors). While Chip-seq data for most of ARFs are still unavailable, prediction of potential AuxRE is restricted by consensus models that detect too many false positive sites. Using sequence analysis of experimentally proven AuxREs, we revealed both an extended nucleotide context pattern for AuxRE itself and three distinct types of its coupling motifs (Y-patch, AuxRE-like, and ABRE-like), which together with AuxRE may form the composite elements. Computational analysis of the genome-wide distribution of the predicted AuxREs and their impact on auxin responsive gene expression allowed us to conclude that: (1) AuxREs are enriched around the transcription start site with the maximum density in 5'UTR; (2) AuxREs mediate auxin responsive up-regulation, not down-regulation. (3) Directly oriented single AuxREs and reverse multiple AuxREs are mostly associated with auxin responsiveness. In the composite AuxRE elements associated with auxin response, ABRE-like and Y-patch are 5'-flanking or overlapping AuxRE, whereas AuxRE-like motif is 3'-flanking. The specificity in location and orientation of the coupling elements suggests them as potential binding sites for ARFs partners.
Figures


Similar articles
-
Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription.BMC Plant Biol. 2012 Aug 1;12:125. doi: 10.1186/1471-2229-12-125. BMC Plant Biol. 2012. PMID: 22852874 Free PMC article.
-
Meta-analysis of transcriptome data identified TGTCNN motif variants associated with the response to plant hormone auxin in Arabidopsis thaliana L.J Bioinform Comput Biol. 2016 Apr;14(2):1641009. doi: 10.1142/S0219720016410092. J Bioinform Comput Biol. 2016. PMID: 27122321
-
Prediction of auxin response elements based on data fusion in Arabidopsis thaliana.Mol Biol Rep. 2018 Oct;45(5):763-772. doi: 10.1007/s11033-018-4216-6. Epub 2018 Jun 23. Mol Biol Rep. 2018. PMID: 29936576
-
Diversity of cis-regulatory elements associated with auxin response in Arabidopsis thaliana.J Exp Bot. 2018 Jan 4;69(2):329-339. doi: 10.1093/jxb/erx254. J Exp Bot. 2018. PMID: 28992117 Free PMC article. Review.
-
Auxin response factors.Curr Opin Plant Biol. 2007 Oct;10(5):453-60. doi: 10.1016/j.pbi.2007.08.014. Epub 2007 Sep 27. Curr Opin Plant Biol. 2007. PMID: 17900969 Review.
Cited by
-
Characterization of Transcription Regulatory Domains of OsMADS29: Identification of Proximal Auxin-Responsive Domains and a Strong Distal Negative Element.Front Plant Sci. 2022 Apr 25;13:850956. doi: 10.3389/fpls.2022.850956. eCollection 2022. Front Plant Sci. 2022. PMID: 35557721 Free PMC article.
-
It's Morphin' time: how multiple signals converge on ARF transcription factors to direct development.Curr Opin Plant Biol. 2020 Oct;57:1-7. doi: 10.1016/j.pbi.2020.04.008. Epub 2020 May 29. Curr Opin Plant Biol. 2020. PMID: 32480312 Free PMC article. Review.
-
metaRE R Package for Meta-Analysis of Transcriptome Data to Identify the cis-Regulatory Code behind the Transcriptional Reprogramming.Genes (Basel). 2020 Jun 9;11(6):634. doi: 10.3390/genes11060634. Genes (Basel). 2020. PMID: 32526881 Free PMC article.
-
Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress.Proc Natl Acad Sci U S A. 2018 Nov 20;115(47):E11178-E11187. doi: 10.1073/pnas.1811491115. Epub 2018 Nov 5. Proc Natl Acad Sci U S A. 2018. PMID: 30397148 Free PMC article.
-
Transcriptional Tuning: How Auxin Strikes Unique Chords in Gene Regulation.Physiol Plant. 2025 May-Jun;177(3):e70229. doi: 10.1111/ppl.70229. Physiol Plant. 2025. PMID: 40302163 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

