Forced unraveling of chromatin fibers with nonuniform linker DNA lengths
- PMID: 25564319
- PMCID: PMC4554754
- DOI: 10.1088/0953-8984/27/6/064113
Forced unraveling of chromatin fibers with nonuniform linker DNA lengths
Abstract
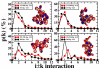
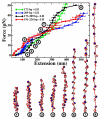
The chromatin fiber undergoes significant structural changes during the cell's life cycle to modulate DNA accessibility. Detailed mechanisms of such structural transformations of chromatin fibers as affected by various internal and external conditions such as the ionic conditions of the medium, the linker DNA length, and the presence of linker histones, constitute an open challenge. Here we utilize Monte Carlo (MC) simulations of a coarse grained model of chromatin with nonuniform linker DNA lengths as found in vivo to help explain some aspects of this challenge. We investigate the unfolding mechanisms of chromatin fibers with alternating linker lengths of 26-62 bp and 44-79 bp using a series of end-to-end stretching trajectories with and without linker histones and compare results to uniform-linker-length fibers. We find that linker histones increase overall resistance of nonuniform fibers and lead to fiber unfolding with superbeads-on-a-string cluster transitions. Chromatin fibers with nonuniform linker DNA lengths display a more complex, multi-step yet smoother process of unfolding compared to their uniform counterparts, likely due to the existence of a more continuous range of nucleosome-nucleosome interactions. This finding echoes the theme that some heterogeneity in fiber component is biologically advantageous.
Figures




References
-
- Maeshima K, Hihara S, Eltsov M. Curr. Opin. Cell. Biol. 2010;22:291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
