Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion
- PMID: 25564401
- PMCID: PMC4351505
- DOI: 10.1182/blood-2014-11-610816
Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion
Abstract
Familial hemophagocytic lymphohistiocytosis (F-HLH) and Griscelli syndrome type 2 (GS) are life-threatening immunodeficiencies characterized by impaired cytotoxic T lymphocyte (CTL) and natural killer (NK) cell lytic activity. In the majority of cases, these disorders are caused by biallelic inactivating germline mutations in genes such as RAB27A (GS) and PRF1, UNC13D, STX11, and STXBP2 (F-HLH). Although monoallelic (ie, heterozygous) mutations have been identified in certain patients, the clinical significance and molecular mechanisms by which these mutations influence CTL and NK cell function remain poorly understood. Here, we characterize 2 novel monoallelic hemophagocytic lymphohistiocytosis (HLH)-associated mutations affecting codon 65 of STXPB2, the gene encoding Munc18-2, a member of the SEC/MUNC18 family. Unlike previously described Munc18-2 mutants, Munc18-2(R65Q) and Munc18-2(R65W) retain the ability to interact with and stabilize syntaxin 11. However, presence of Munc18-2(R65Q/W) in patient-derived lymphocytes and forced expression in control CTLs and NK cells diminishes degranulation and cytotoxic activity. Mechanistic studies reveal that mutations affecting R65 hinder membrane fusion in vitro by arresting the late steps of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-complex assembly. Collectively, these results reveal a direct role for SEC/MUNC18 proteins in promoting SNARE-complex assembly in vivo and suggest that STXBP2 R65 mutations operate in a novel dominant-negative fashion to impair lytic granule fusion and contribute to HLH.
© 2015 by The American Society of Hematology.
Figures
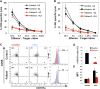


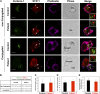



Similar articles
-
SM protein Munc18-2 facilitates transition of Syntaxin 11-mediated lipid mixing to complete fusion for T-lymphocyte cytotoxicity.Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):E2176-E2185. doi: 10.1073/pnas.1617981114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265073 Free PMC article.
-
STXBP2-R190C Variant in a Patient With Neonatal Hemophagocytic Lymphohistiocytosis (HLH) and G6PD Deficiency Reveals a Critical Role of STXBP2 Domain 2 on Granule Exocytosis.Front Immunol. 2020 Oct 8;11:545414. doi: 10.3389/fimmu.2020.545414. eCollection 2020. Front Immunol. 2020. PMID: 33162974 Free PMC article.
-
Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells.J Clin Invest. 2009 Dec;119(12):3765-73. doi: 10.1172/JCI40732. Epub 2009 Nov 2. J Clin Invest. 2009. PMID: 19884660 Free PMC article.
-
A unique SNARE machinery for exocytosis of cytotoxic granules and platelets granules.Mol Membr Biol. 2015;32(4):120-6. doi: 10.3109/09687688.2015.1079934. Mol Membr Biol. 2015. PMID: 26508555 Review.
-
Angeborene hämophagozytische Lymphohistiozytose (HLH).Klin Padiatr. 2010 Nov;222(6):345-50. doi: 10.1055/s-0029-1246165. Epub 2010 May 10. Klin Padiatr. 2010. PMID: 20458667 Review.
Cited by
-
The septin cytoskeleton regulates natural killer cell lytic granule release.J Cell Biol. 2020 Nov 2;219(11):e202002145. doi: 10.1083/jcb.202002145. J Cell Biol. 2020. PMID: 32841357 Free PMC article.
-
SM protein Munc18-2 facilitates transition of Syntaxin 11-mediated lipid mixing to complete fusion for T-lymphocyte cytotoxicity.Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):E2176-E2185. doi: 10.1073/pnas.1617981114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265073 Free PMC article.
-
Breaking Down Barriers: Epithelial Contributors to Monogenic IBD Pathogenesis.Inflamm Bowel Dis. 2024 Jul 3;30(7):1189-1206. doi: 10.1093/ibd/izad319. Inflamm Bowel Dis. 2024. PMID: 38280053 Free PMC article. Review.
-
A collection of patient-derived intestinal organoid lines reveals epithelial phenotypes associated with genetic drivers of pediatric inflammatory bowel disease.bioRxiv [Preprint]. 2025 Jun 16:2025.06.11.659052. doi: 10.1101/2025.06.11.659052. bioRxiv. 2025. PMID: 40666956 Free PMC article. Preprint.
-
Bacteria-Associated Cytokine Storm Syndrome.Adv Exp Med Biol. 2024;1448:275-283. doi: 10.1007/978-3-031-59815-9_19. Adv Exp Med Biol. 2024. PMID: 39117821 Review.
References
-
- Stepp SE, Dufourcq-Lagelouse R, Le Deist F, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286(5446):1957–1959. - PubMed
-
- Feldmann J, Callebaut I, Raposo G, et al. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3). Cell. 2003;115(4):461–473. - PubMed
-
- zur Stadt U, Schmidt S, Kasper B, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet. 2005;14(6):827–834. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

