Collaboration between grass seedlings and rhizobacteria to scavenge organic nitrogen in soils
- PMID: 25564515
- PMCID: PMC4313791
- DOI: 10.1093/aobpla/plu093
Collaboration between grass seedlings and rhizobacteria to scavenge organic nitrogen in soils
Abstract
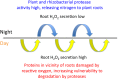
Plants require nitrogen (N) to make proteins, nucleic acids and other biological molecules. It is widely accepted that plants absorb inorganic forms of N to fill their needs. However, recently it has become clear that plants also have the capacity to absorb organic N from soils. In this paper we describe a new kind of symbiosis involving seed-vectored rhizobacteria and grasses that is targeted at enhancing acquisition of organic N from soils. Our proposal is based on results of experiments on seedlings of grass species Festuca arundinacea Schreb., Lolium perenne L. and Poa annua L. that suggest: (i) seed-vectored rhizobacteria colonize seedling roots and influence their development; (ii) reactive oxygen secretion by seedling roots plays a role in organic N procurement by denaturing microbial proteins in the vicinity of roots (daytime activity); and (iii) plant root and microbial proteases degrade denatured proteins prior to absorption by roots (night-time activity). This research involved the following types of studies: (i) seedling root development experiments with and without rhizobacteria on a variety of substrates in agarose media and (ii) isotopic N-tracking experiments to evaluate the absorption into seedlings of N obtained from degradation of proteins. We hypothesize that grasses, in particular, are adapted to scavenge organic N from soils through application of this 'oxidative nitrogen scavenging' symbiosis with rhizobacteria, and their soil-permeating root systems. This newly discovered symbiosis in grass species could lead to new ways to cultivate and manage grasses to enhance efficiency of N utilization and reduce applications of inorganic fertilizers.
Keywords: Grasses; microbiome; nitrogen use efficiency; oxidative nitrogen scavenging; plant growth-promoting rhizobacteria; symbiosis.
Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures




References
-
- Bott R, Ultsch M, Kossiakoff A, Grayear T, Katz B, Power S. The three-dimensional structure of Bacillus amyloliquefaciens subtilisin at 1.8 Å and an analysis of the structural consequences of peroxide inactivation. The Journal of Biological Chemistry. 1988;263:7895–7906. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

